Search Results for author: Henning Muller
Found 7 papers, 5 papers with code
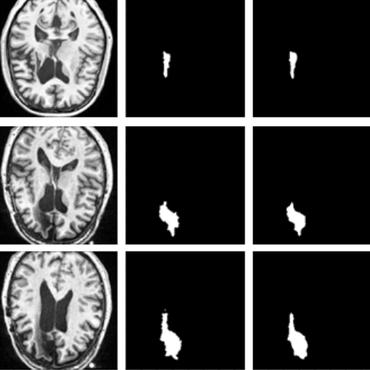
Structural-Based Uncertainty in Deep Learning Across Anatomical Scales: Analysis in White Matter Lesion Segmentation
1 code implementation • 15 Nov 2023 • Nataliia Molchanova, Vatsal Raina, Andrey Malinin, Francesco La Rosa, Adrien Depeursinge, Mark Gales, Cristina Granziera, Henning Muller, Mara Graziani, Meritxell Bach Cuadra
The results from a multi-centric MRI dataset of 172 patients demonstrate that our proposed measures more effectively capture model errors at the lesion and patient scales compared to measures that average voxel-scale uncertainty values.
Disentangling Neuron Representations with Concept Vectors
1 code implementation • 19 Apr 2023 • Laura O'Mahony, Vincent Andrearczyk, Henning Muller, Mara Graziani
Mechanistic interpretability aims to understand how models store representations by breaking down neural networks into interpretable units.
Tackling Bias in the Dice Similarity Coefficient: Introducing nDSC for White Matter Lesion Segmentation
1 code implementation • 10 Feb 2023 • Vatsal Raina, Nataliia Molchanova, Mara Graziani, Andrey Malinin, Henning Muller, Meritxell Bach Cuadra, Mark Gales
This work describes a detailed analysis of the recently proposed normalised Dice Similarity Coefficient (nDSC) for binary segmentation tasks as an adaptation of DSC which scales the precision at a fixed recall rate to tackle this bias.
Unsupervised Method for Intra-patient Registration of Brain Magnetic Resonance Images based on Objective Function Weighting by Inverse Consistency: Contribution to the BraTS-Reg Challenge
no code implementations • 14 Nov 2022 • Marek Wodzinski, Artur Jurgas, Niccolo Marini, Manfredo Atzori, Henning Muller
The importance of this task motivated researchers to organize the BraTS-Reg challenge, jointly with IEEE ISBI 2022 and MICCAI 2022 conferences.
Novel structural-scale uncertainty measures and error retention curves: application to multiple sclerosis
1 code implementation • 9 Nov 2022 • Nataliia Molchanova, Vatsal Raina, Andrey Malinin, Francesco La Rosa, Henning Muller, Mark Gales, Cristina Granziera, Mara Graziani, Meritxell Bach Cuadra
This paper focuses on the uncertainty estimation for white matter lesions (WML) segmentation in magnetic resonance imaging (MRI).
The Brain Tumor Sequence Registration (BraTS-Reg) Challenge: Establishing Correspondence Between Pre-Operative and Follow-up MRI Scans of Diffuse Glioma Patients
no code implementations • 13 Dec 2021 • Bhakti Baheti, Satrajit Chakrabarty, Hamed Akbari, Michel Bilello, Benedikt Wiestler, Julian Schwarting, Evan Calabrese, Jeffrey Rudie, Syed Abidi, Mina Mousa, Javier Villanueva-Meyer, Brandon K. K. Fields, Florian Kofler, Russell Takeshi Shinohara, Juan Eugenio Iglesias, Tony C. W. Mok, Albert C. S. Chung, Marek Wodzinski, Artur Jurgas, Niccolo Marini, Manfredo Atzori, Henning Muller, Christoph Grobroehmer, Hanna Siebert, Lasse Hansen, Mattias P. Heinrich, Luca Canalini, Jan Klein, Annika Gerken, Stefan Heldmann, Alessa Hering, Horst K. Hahn, Mingyuan Meng, Lei Bi, Dagan Feng, Jinman Kim, Ramy A. Zeineldin, Mohamed E. Karar, Franziska Mathis-Ullrich, Oliver Burgert, Javid Abderezaei, Aymeric Pionteck, Agamdeep Chopra, Mehmet Kurt, Kewei Yan, Yonghong Yan, Zhe Tang, Jianqiang Ma, Sahar Almahfouz Nasser, Nikhil Cherian Kurian, Mohit Meena, Saqib Shamsi, Amit Sethi, Nicholas J. Tustison, Brian B. Avants, Philip Cook, James C. Gee, Lin Tian, Hastings Greer, Marc Niethammer, Andrew Hoopes, Malte Hoffmann, Adrian V. Dalca, Stergios Christodoulidis, Theo Estiene, Maria Vakalopoulou, Nikos Paragios, Daniel S. Marcus, Christos Davatzikos, Aristeidis Sotiras, Bjoern Menze, Spyridon Bakas, Diana Waldmannstetter
Registration of longitudinal brain MRI scans containing pathologies is challenging due to dramatic changes in tissue appearance.
Learning Interpretable Microscopic Features of Tumor by Multi-task Adversarial CNNs To Improve Generalization
1 code implementation • 4 Aug 2020 • Mara Graziani, Sebastian Otalora, Stephane Marchand-Maillet, Henning Muller, Vincent Andrearczyk
Here we show that our architecture, by learning end-to-end an uncertainty-based weighting combination of multi-task and adversarial losses, is encouraged to focus on pathology features such as density and pleomorphism of nuclei, e. g. variations in size and appearance, while discarding misleading features such as staining differences.