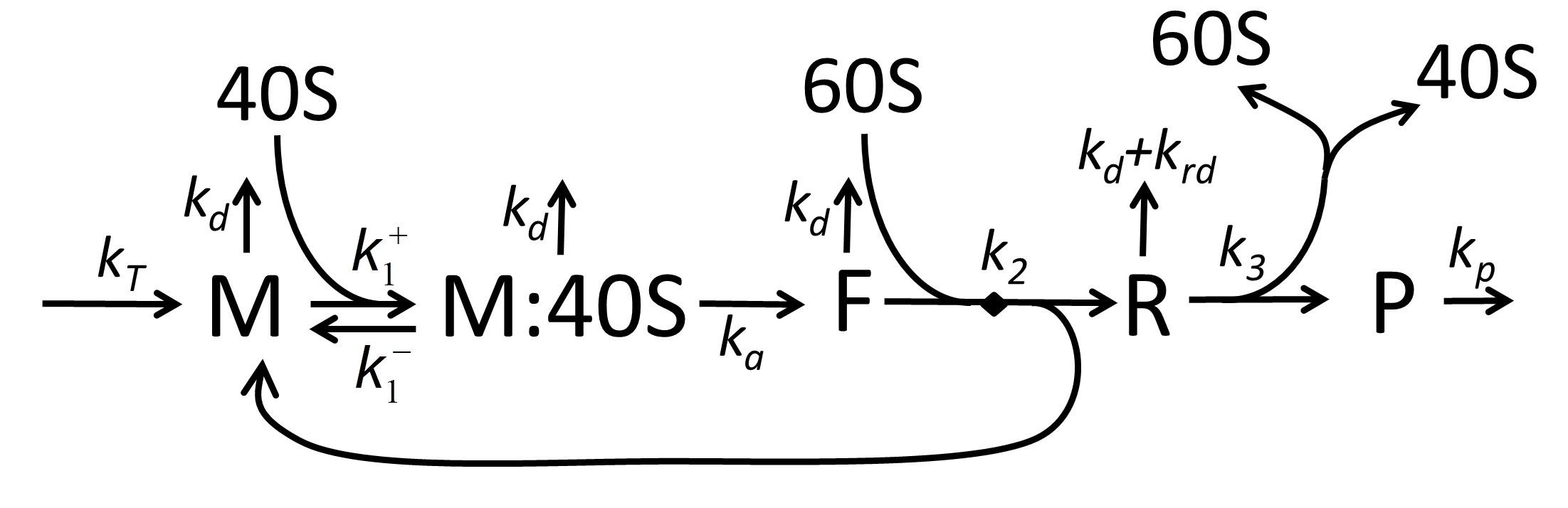
Basic and simple mathematical model of coupled transcription, translation and degradation
Synthesis of proteins is one of the most fundamental biological processes, which consumes a significant amount of cellular resources. Despite many efforts to produce detailed mechanistic mathematical models of translation, no basic and simple kinetic model of mRNA lifecycle (transcription, translation and degradation) exists. We build such a model by lumping multiple states of translated mRNA into few dynamical variables and introducing a pool of translating ribosomes. The basic and simple model can be extended, if necessary, to take into account various phenomena such as the interaction between translating ribosomes or regulation of translation by microRNA. The model can be used as a building block (translation module) for more complex models of cellular processes.
PDF Abstract