3D Mitochondria Instance Segmentation with Spatio-Temporal Transformers
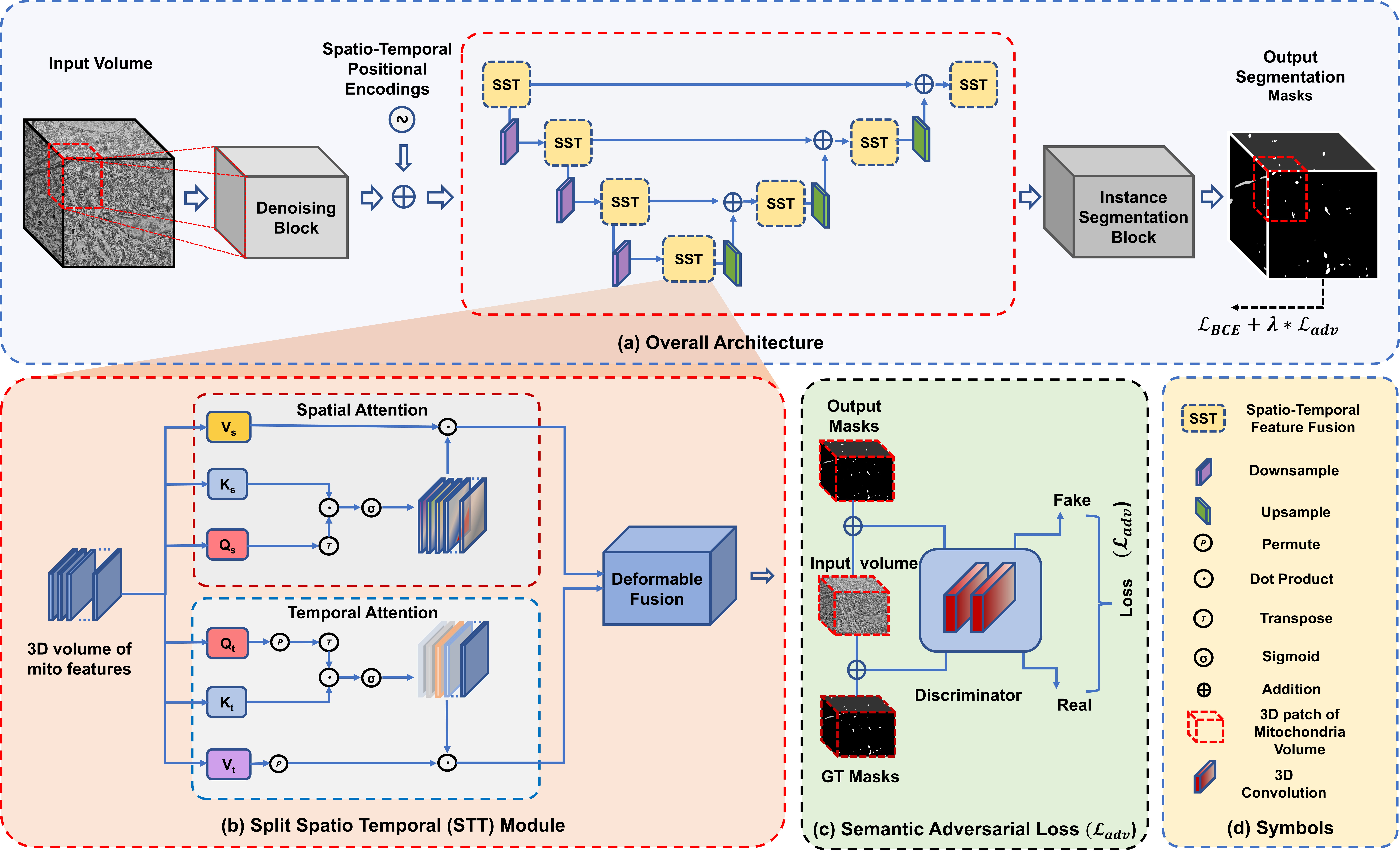
Accurate 3D mitochondria instance segmentation in electron microscopy (EM) is a challenging problem and serves as a prerequisite to empirically analyze their distributions and morphology. Most existing approaches employ 3D convolutions to obtain representative features. However, these convolution-based approaches struggle to effectively capture long-range dependencies in the volume mitochondria data, due to their limited local receptive field. To address this, we propose a hybrid encoder-decoder framework based on a split spatio-temporal attention module that efficiently computes spatial and temporal self-attentions in parallel, which are later fused through a deformable convolution. Further, we introduce a semantic foreground-background adversarial loss during training that aids in delineating the region of mitochondria instances from the background clutter. Our extensive experiments on three benchmarks, Lucchi, MitoEM-R and MitoEM-H, reveal the benefits of the proposed contributions achieving state-of-the-art results on all three datasets. Our code and models are available at https://github.com/OmkarThawakar/STT-UNET.
PDF Abstract