Enhancing Clinical BERT Embedding using a Biomedical Knowledge Base
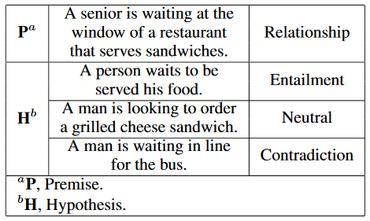
Domain knowledge is important for building Natural Language Processing (NLP) systems for low-resource settings, such as in the clinical domain. In this paper, a novel joint training method is introduced for adding knowledge base information from the Unified Medical Language System (UMLS) into language model pre-training for some clinical domain corpus. We show that in three different downstream clinical NLP tasks, our pre-trained language model outperforms the corresponding model with no knowledge base information and other state-of-the-art models. Specifically, in a natural language inference task applied to clinical texts, our knowledge base pre-training approach improves accuracy by up to 1.7{\%}, whereas in clinical name entity recognition tasks, the F1-score improves by up to 1.0{\%}. The pre-trained models are available at https://github.com/noc-lab/clinical-kb-bert.
PDF Abstract


 MIMIC-III
MIMIC-III