A computational approach to aid clinicians in selecting anti-viral drugs for COVID-19 trials
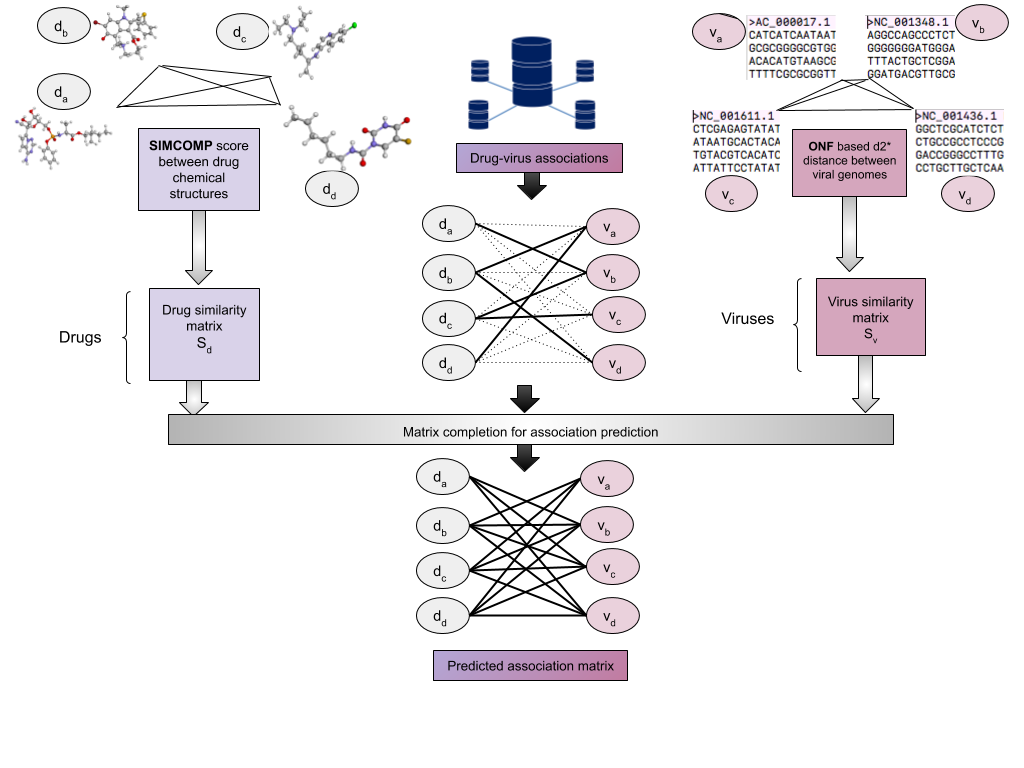
COVID-19 has fast-paced drug re-positioning for its treatment. This work builds computational models for the same. The aim is to assist clinicians with a tool for selecting prospective antiviral treatments. Since the virus is known to mutate fast, the tool is likely to help clinicians in selecting the right set of antivirals for the mutated isolate. The main contribution of this work is a manually curated database publicly shared, comprising of existing associations between viruses and their corresponding antivirals. The database gathers similarity information using the chemical structure of drugs and the genomic structure of viruses. Along with this database, we make available a set of state-of-the-art computational drug re-positioning tools based on matrix completion. The tools are first analysed on a standard set of experimental protocols for drug target interactions. The best performing ones are applied for the task of re-positioning antivirals for COVID-19. These tools select six drugs out of which four are currently under various stages of trial, namely Remdesivir (as a cure), Ribavarin (in combination with others for cure), Umifenovir (as a prophylactic and cure) and Sofosbuvir (as a cure). Another unanimous prediction is Tenofovir alafenamide, which is a novel tenofovir prodrug developed in order to improve renal safety when compared to the counterpart tenofovir disoproxil. Both are under trail, the former as a cure and the latter as a prophylactic. These results establish that the computational methods are in sync with the state-of-practice. We also demonstrate how the selected drugs change as the SARS-Cov-2 mutates over time, suggesting the importance of such a tool in drug prediction. The dataset and software is available publicly at https://github.com/aanchalMongia/DVA and the prediction tool with a user-friendly interface is available at http://dva.salsa.iiitd.edu.in.
PDF Abstract