A Hebbian/Anti-Hebbian Network Derived from Online Non-Negative Matrix Factorization Can Cluster and Discover Sparse Features
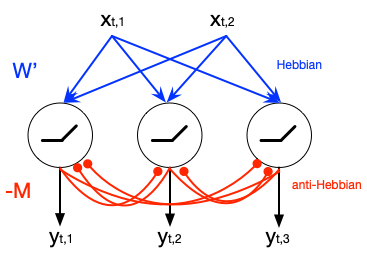
Despite our extensive knowledge of biophysical properties of neurons, there is no commonly accepted algorithmic theory of neuronal function. Here we explore the hypothesis that single-layer neuronal networks perform online symmetric nonnegative matrix factorization (SNMF) of the similarity matrix of the streamed data. By starting with the SNMF cost function we derive an online algorithm, which can be implemented by a biologically plausible network with local learning rules. We demonstrate that such network performs soft clustering of the data as well as sparse feature discovery. The derived algorithm replicates many known aspects of sensory anatomy and biophysical properties of neurons including unipolar nature of neuronal activity and synaptic weights, local synaptic plasticity rules and the dependence of learning rate on cumulative neuronal activity. Thus, we make a step towards an algorithmic theory of neuronal function, which should facilitate large-scale neural circuit simulations and biologically inspired artificial intelligence.
PDF Abstract