A novel algorithmic approach to Bayesian Logic Regression
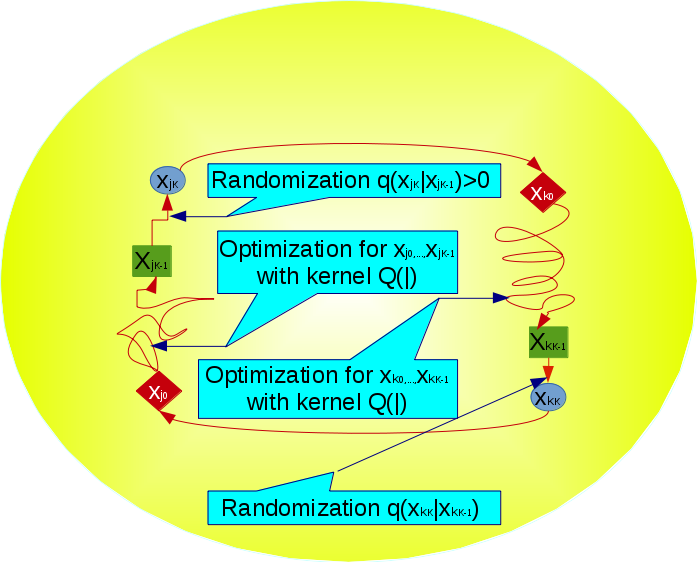
Logic regression was developed more than a decade ago as a tool to construct predictors from Boolean combinations of binary covariates. It has been mainly used to model epistatic effects in genetic association studies, which is very appealing due to the intuitive interpretation of logic expressions to describe the interaction between genetic variations. Nevertheless logic regression has remained less well known than other approaches to epistatic association mapping. Here we will adopt an advanced evolutionary algorithm called GMJMCMC (Genetically modified Mode Jumping Markov Chain Monte Carlo) to perform Bayesian model selection in the space of logic regression models. After describing the algorithmic details of GMJMCMC we perform a comprehensive simulation study that illustrates its performance given logic regression terms of various complexity. Specifically GMJMCMC is shown to be able to identify three-way and even four-way interactions with relatively large power, a level of complexity which has not been achieved by previous implementations of logic regression. We apply GMJMCMC to reanalyze QTL mapping data for Recombinant Inbred Lines in Arabidopsis thaliana and from a backcross population in Drosophila where we identify several interesting epistatic effects
PDF Abstract