Automated assessment of disease severity of COVID-19 using artificial intelligence with synthetic chest CT
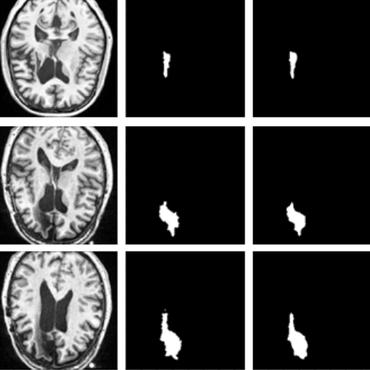
Background: Triage of patients is important to control the pandemic of coronavirus disease 2019 (COVID-19), especially during the peak of the pandemic when clinical resources become extremely limited. Purpose: To develop a method that automatically segments and quantifies lung and pneumonia lesions with synthetic chest CT and assess disease severity in COVID-19 patients. Materials and Methods: In this study, we incorporated data augmentation to generate synthetic chest CT images using public available datasets (285 datasets from "Lung Nodule Analysis 2016"). The synthetic images and masks were used to train a 2D U-net neural network and tested on 203 COVID-19 datasets to generate lung and lesion segmentations. Disease severity scores (DL: damage load; DS: damage score) were calculated based on the segmentations. Correlations between DL/DS and clinical lab tests were evaluated using Pearson's method. A p-value < 0.05 was considered as statistical significant. Results: Automatic lung and lesion segmentations were compared with manual annotations. For lung segmentation, the median values of dice similarity coefficient, Jaccard index and average surface distance, were 98.56%, 97.15% and 0.49 mm, respectively. The same metrics for lesion segmentation were 76.95%, 62.54% and 2.36 mm, respectively. Significant (p << 0.05) correlations were found between DL/DS and percentage lymphocytes tests, with r-values of -0.561 and -0.501, respectively. Conclusion: An AI system that based on thoracic radiographic and data augmentation was proposed to segment lung and lesions in COVID-19 patients. Correlations between imaging findings and clinical lab tests suggested the value of this system as a potential tool to assess disease severity of COVID-19.
PDF Abstract