BioNLI: Generating a Biomedical NLI Dataset Using Lexico-semantic Constraints for Adversarial Examples
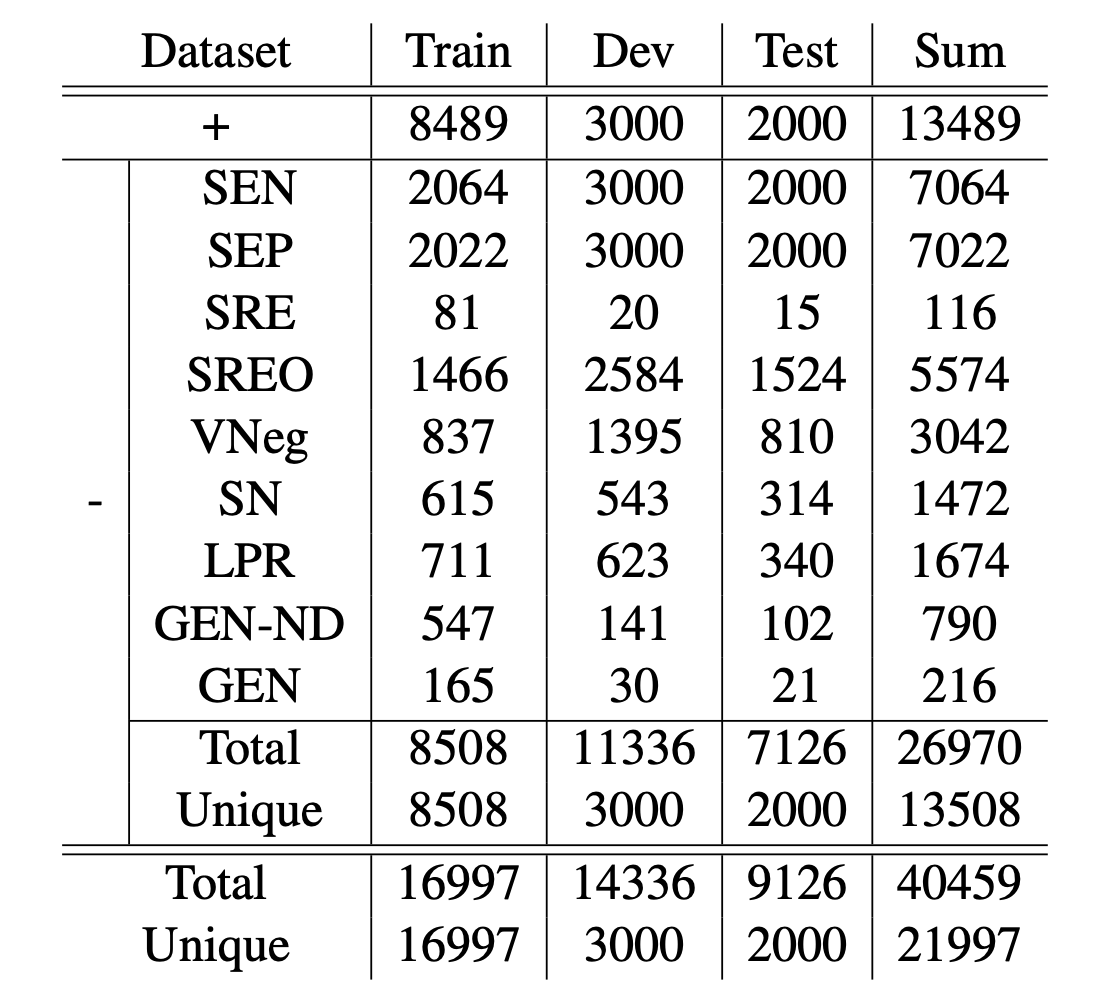
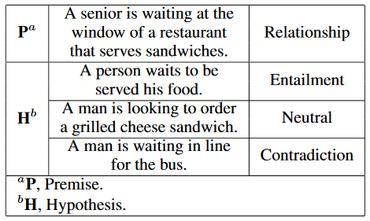
Natural language inference (NLI) is critical for complex decision-making in biomedical domain. One key question, for example, is whether a given biomedical mechanism is supported by experimental evidence. This can be seen as an NLI problem but there are no directly usable datasets to address this. The main challenge is that manually creating informative negative examples for this task is difficult and expensive. We introduce a novel semi-supervised procedure that bootstraps an NLI dataset from existing biomedical dataset that pairs mechanisms with experimental evidence in abstracts. We generate a range of negative examples using nine strategies that manipulate the structure of the underlying mechanisms both with rules, e.g., flip the roles of the entities in the interaction, and, more importantly, as perturbations via logical constraints in a neuro-logical decoding system. We use this procedure to create a novel dataset for NLI in the biomedical domain, called BioNLI and benchmark two state-of-the-art biomedical classifiers. The best result we obtain is around mid 70s in F1, suggesting the difficulty of the task. Critically, the performance on the different classes of negative examples varies widely, from 97% F1 on the simple role change negative examples, to barely better than chance on the negative examples generated using neuro-logic decoding.
PDF Abstract| Task | Dataset | Model | Metric Name | Metric Value | Global Rank | Benchmark |
|---|---|---|---|---|---|---|
| Natural Language Inference | BioNLI | BioLinkBert | Macro F1 | 0.77 | # 1 |



 BioNLI
BioNLI
