Contrastive learning-based computational histopathology predict differential expression of cancer driver genes
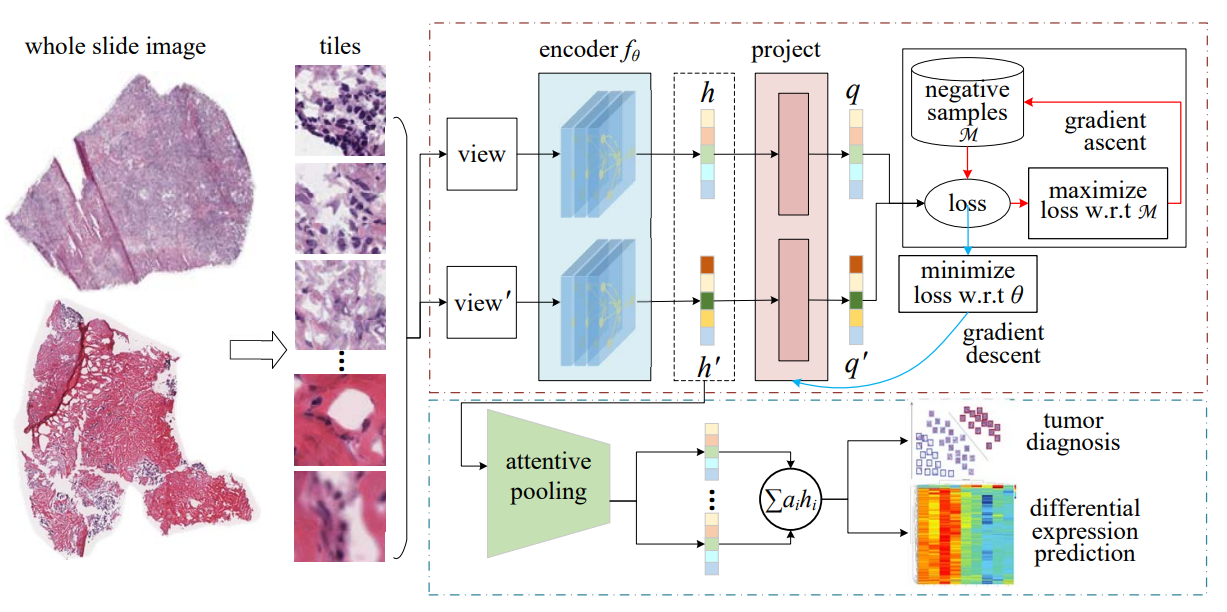
Digital pathological analysis is run as the main examination used for cancer diagnosis. Recently, deep learning-driven feature extraction from pathology images is able to detect genetic variations and tumor environment, but few studies focus on differential gene expression in tumor cells. In this paper, we propose a self-supervised contrastive learning framework, HistCode, to infer differential gene expressions from whole slide images (WSIs). We leveraged contrastive learning on large-scale unannotated WSIs to derive slide-level histopathological feature in latent space, and then transfer it to tumor diagnosis and prediction of differentially expressed cancer driver genes. Our extensive experiments showed that our method outperformed other state-of-the-art models in tumor diagnosis tasks, and also effectively predicted differential gene expressions. Interestingly, we found the higher fold-changed genes can be more precisely predicted. To intuitively illustrate the ability to extract informative features from pathological images, we spatially visualized the WSIs colored by the attentive scores of image tiles. We found that the tumor and necrosis areas were highly consistent with the annotations of experienced pathologists. Moreover, the spatial heatmap generated by lymphocyte-specific gene expression patterns was also consistent with the manually labeled WSI.
PDF Abstract