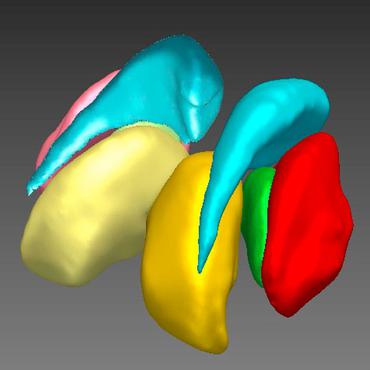
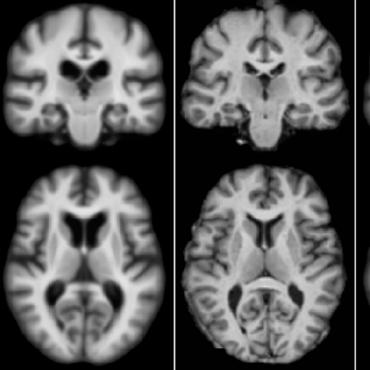
Direct cortical thickness estimation using deep learning‐based anatomy segmentation and cortex parcellation
Accurate and reliable measures of cortical thickness from magnetic resonance imaging are an important biomarker to study neurodegenerative and neurological disorders. Diffeomorphic registration‐based cortical thickness (DiReCT) is a known technique to derive such measures from non‐surface‐based volumetric tissue maps. ANTs provides an open‐source method for estimating cortical thickness, derived by applying DiReCT to an atlas‐based segmentation. In this paper, we propose DL+DiReCT, a method using high‐quality deep learning‐based neuroanatomy segmentations followed by DiReCT, yielding accurate and reliable cortical thickness measures in a short time. We evaluate the methods on two independent datasets and compare the results against surface‐based measures from FreeSurfer. Good correlation of DL+DiReCT with FreeSurfer was observed (r = .887) for global mean cortical thickness compared to ANTs versus FreeSurfer (r = .608). Experiments suggest that both DiReCT‐based methods had higher sensitivity to changes in cortical thickness than Freesurfer. However, while ANTs showed low scan‐rescan robustness, DL+DiReCT showed similar robustness to Freesurfer. Effect‐sizes for group‐wise differences of healthy controls compared to individuals with dementia were highest with the deep learning‐based segmentation. DL+DiReCT is a promising combination of a deep learning‐based method with a traditional registration technique to detect subtle changes in cortical thickness.
PDF Abstract