DPPIN: A Biological Repository of Dynamic Protein-Protein Interaction Network Data
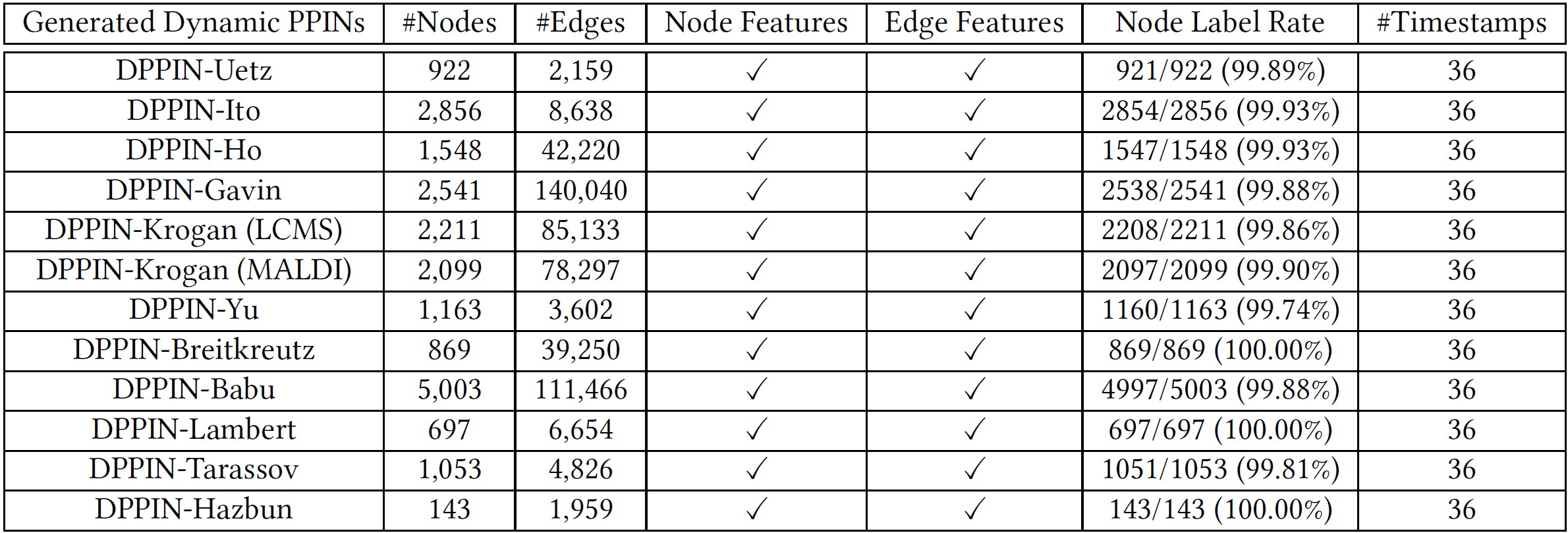
In the big data era, the relationship between entries becomes more and more complex. Many graph (or network) algorithms have already paid attention to dynamic networks, which are more suitable than static ones for fitting the complex real-world scenarios with evolving structures and features. To contribute to the dynamic network representation learning and mining research, we provide a new bunch of label-adequate, dynamics-meaningful, and attribute-sufficient dynamic networks from the health domain. To be specific, in our proposed repository DPPIN, we totally have 12 individual dynamic network datasets at different scales, and each dataset is a dynamic protein-protein interaction network describing protein-level interactions of yeast cells. We hope these domain-specific node features, structure evolution patterns, and node and graph labels could inspire the regularization techniques to increase the performance of graph machine learning algorithms in a more complex setting. Also, we link potential applications with our DPPIN by designing various dynamic graph experiments, where DPPIN could indicate future research opportunities for some tasks by presenting challenges on state-of-the-art baseline algorithms. Finally, we identify future directions to improve the utility of this repository and welcome constructive inputs from the community. All resources (e.g., data and code) of this work are deployed and publicly available at https://github.com/DongqiFu/DPPIN.
PDF AbstractCode
Datasets
Introduced in the Paper:
 DPPIN
DPPIN