Drug and Disease Interpretation Learning with Biomedical Entity Representation Transformer
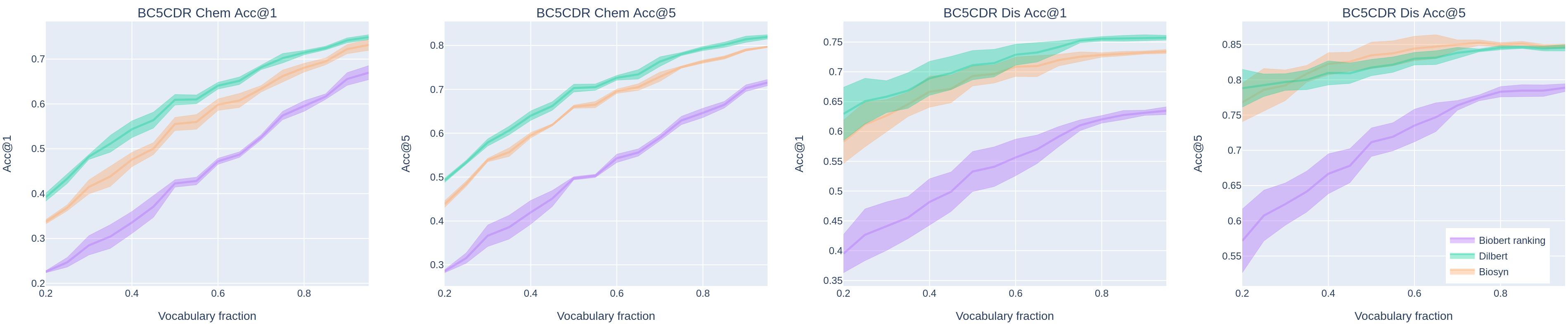
Concept normalization in free-form texts is a crucial step in every text-mining pipeline. Neural architectures based on Bidirectional Encoder Representations from Transformers (BERT) have achieved state-of-the-art results in the biomedical domain. In the context of drug discovery and development, clinical trials are necessary to establish the efficacy and safety of drugs. We investigate the effectiveness of transferring concept normalization from the general biomedical domain to the clinical trials domain in a zero-shot setting with an absence of labeled data. We propose a simple and effective two-stage neural approach based on fine-tuned BERT architectures. In the first stage, we train a metric learning model that optimizes relative similarity of mentions and concepts via triplet loss. The model is trained on available labeled corpora of scientific abstracts to obtain vector embeddings of concept names and entity mentions from texts. In the second stage, we find the closest concept name representation in an embedding space to a given clinical mention. We evaluated several models, including state-of-the-art architectures, on a dataset of abstracts and a real-world dataset of trial records with interventions and conditions mapped to drug and disease terminologies. Extensive experiments validate the effectiveness of our approach in knowledge transfer from the scientific literature to clinical trials.
PDF Abstract