Drug Re-positioning via Text Augmented Knowledge Graph Embeddings
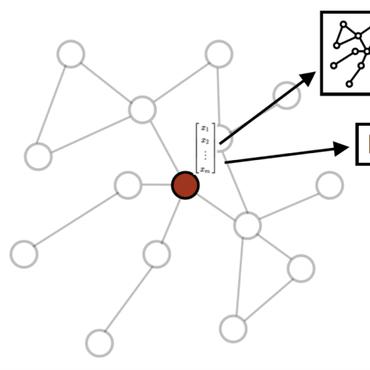
Drug re-positioning, modeled as a link prediction problem over medical knowledge graphs (KG), has great potential in finding new usage or targets for approved medicine with relatively low cost. However, the semantic information in medical KGs is rarely utilized, let alone the external medical databases curated by domain experts. This work attempts to integrate textual descriptions of biomedical KG entities in training knowledge graph embeddings (KGE) and evaluates its effectiveness for drug re-positioning. We implement multiple text augmentation methods on TransE as a case study and further apply the best method on other embedding models. Both qualitative and quantitative error analyses with two novel metrics are conducted to shed light on the effects of adding textual information in our model. We conclude that textual information is generally useful, but it may also backfire.
PDF Abstract