Dynamic Deep Networks for Retinal Vessel Segmentation
Segmenting the retinal vasculature entails a trade-off between how much of the overall vascular structure we identify vs. how precisely we segment individual vessels. In particular, state-of-the-art methods tend to under-segment faint vessels, as well as pixels that lie on the edges of thicker vessels. Thus, they underestimate the width of individual vessels, as well as the ratio of large to small vessels. More generally, many crucial bio-markers---including the artery-vein (AV) ratio, branching angles, number of bifurcation, fractal dimension, tortuosity, vascular length-to-diameter ratio and wall-to-lumen length---require precise measurements of individual vessels. To address this limitation, we propose a novel, stochastic training scheme for deep neural networks that better classifies the faint, ambiguous regions of the image. Our approach relies on two key innovations. First, we train our deep networks with dynamic weights that fluctuate during each training iteration. This stochastic approach forces the network to learn a mapping that robustly balances precision and recall. Second, we decouple the segmentation process into two steps. In the first half of our pipeline, we estimate the likelihood of every pixel and then use these likelihoods to segment pixels that are clearly vessel or background. In the latter part of our pipeline, we use a second network to classify the ambiguous regions in the image. Our proposed method obtained state-of-the-art results on five retinal datasets---DRIVE, STARE, CHASE-DB, AV-WIDE, and VEVIO---by learning a robust balance between false positive and false negative rates. In addition, we are the first to report segmentation results on the AV-WIDE dataset, and we have made the ground-truth annotations for this dataset publicly available.
PDF Abstract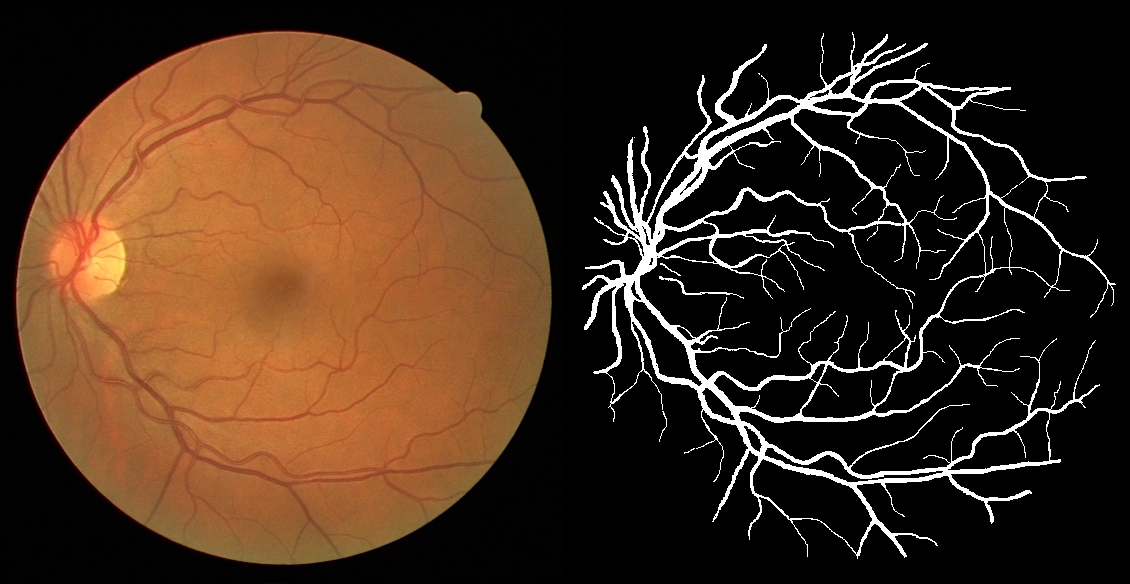


 DRIVE
DRIVE
 STARE
STARE