Ensembling Low Precision Models for Binary Biomedical Image Segmentation
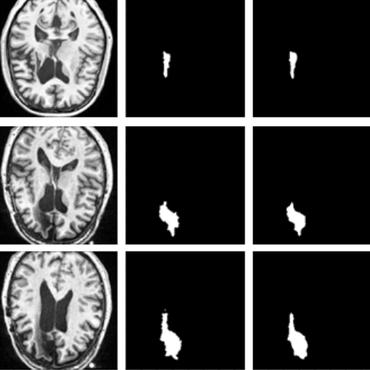
Segmentation of anatomical regions of interest such as vessels or small lesions in medical images is still a difficult problem that is often tackled with manual input by an expert. One of the major challenges for this task is that the appearance of foreground (positive) regions can be similar to background (negative) regions. As a result, many automatic segmentation algorithms tend to exhibit asymmetric errors, typically producing more false positives than false negatives. In this paper, we aim to leverage this asymmetry and train a diverse ensemble of models with very high recall, while sacrificing their precision. Our core idea is straightforward: A diverse ensemble of low precision and high recall models are likely to make different false positive errors (classifying background as foreground in different parts of the image), but the true positives will tend to be consistent. Thus, in aggregate the false positive errors will cancel out, yielding high performance for the ensemble. Our strategy is general and can be applied with any segmentation model. In three different applications (carotid artery segmentation in a neck CT angiography, myocardium segmentation in a cardiovascular MRI and multiple sclerosis lesion segmentation in a brain MRI), we show how the proposed approach can significantly boost the performance of a baseline segmentation method.
PDF Abstract