GeNet: Deep Representations for Metagenomics
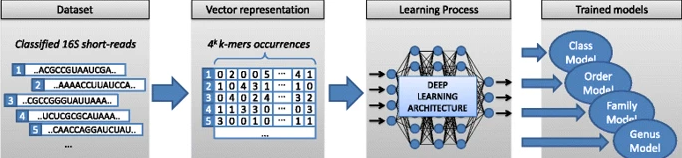
We introduce GeNet, a method for shotgun metagenomic classification from raw DNA sequences that exploits the known hierarchical structure between labels for training. We provide a comparison with state-of-the-art methods Kraken and Centrifuge on datasets obtained from several sequencing technologies, in which dataset shift occurs. We show that GeNet obtains competitive precision and good recall, with orders of magnitude less memory requirements. Moreover, we show that a linear model trained on top of representations learned by GeNet achieves recall comparable to state-of-the-art methods on the aforementioned datasets, and achieves over 90% accuracy in a challenging pathogen detection problem. This provides evidence of the usefulness of the representations learned by GeNet for downstream biological tasks.
PDF Abstract