Graph Attention Networks
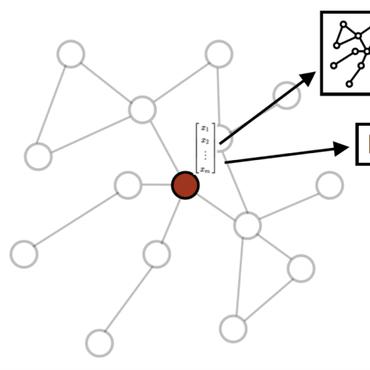
We present graph attention networks (GATs), novel neural network architectures that operate on graph-structured data, leveraging masked self-attentional layers to address the shortcomings of prior methods based on graph convolutions or their approximations. By stacking layers in which nodes are able to attend over their neighborhoods' features, we enable (implicitly) specifying different weights to different nodes in a neighborhood, without requiring any kind of costly matrix operation (such as inversion) or depending on knowing the graph structure upfront. In this way, we address several key challenges of spectral-based graph neural networks simultaneously, and make our model readily applicable to inductive as well as transductive problems. Our GAT models have achieved or matched state-of-the-art results across four established transductive and inductive graph benchmarks: the Cora, Citeseer and Pubmed citation network datasets, as well as a protein-protein interaction dataset (wherein test graphs remain unseen during training).
PDF Abstract ICLR 2018 PDF ICLR 2018 AbstractCode
Datasets
 CIFAR-10
Pubmed
CIFAR-10
Pubmed
 OGB
OGB
 Flickr30k
Flickr30k
 Cora
Citeseer
PPI
Cora
Citeseer
PPI
 JHMDB
JHMDB
 ZINC
Penn94
genius
Deezer-Europe
Wisconsin(60%/20%/20% random splits)
Squirrel (60%/20%/20% random splits)
Film (60%/20%/20% random splits)
Texas(60%/20%/20% random splits)
Cornell (60%/20%/20% random splits)
PubMed (60%/20%/20% random splits)
Chameleon(60%/20%/20% random splits)
USA Air-Traffic
Brazil Air-Traffic
ZINC
Penn94
genius
Deezer-Europe
Wisconsin(60%/20%/20% random splits)
Squirrel (60%/20%/20% random splits)
Film (60%/20%/20% random splits)
Texas(60%/20%/20% random splits)
Cornell (60%/20%/20% random splits)
PubMed (60%/20%/20% random splits)
Chameleon(60%/20%/20% random splits)
USA Air-Traffic
Brazil Air-Traffic
Results from the Paper
 Ranked #1 on
Node Classification
on Pubmed
(Validation metric)
Ranked #1 on
Node Classification
on Pubmed
(Validation metric)