HiGNN: Hierarchical Informative Graph Neural Networks for Molecular Property Prediction Equipped with Feature-Wise Attention
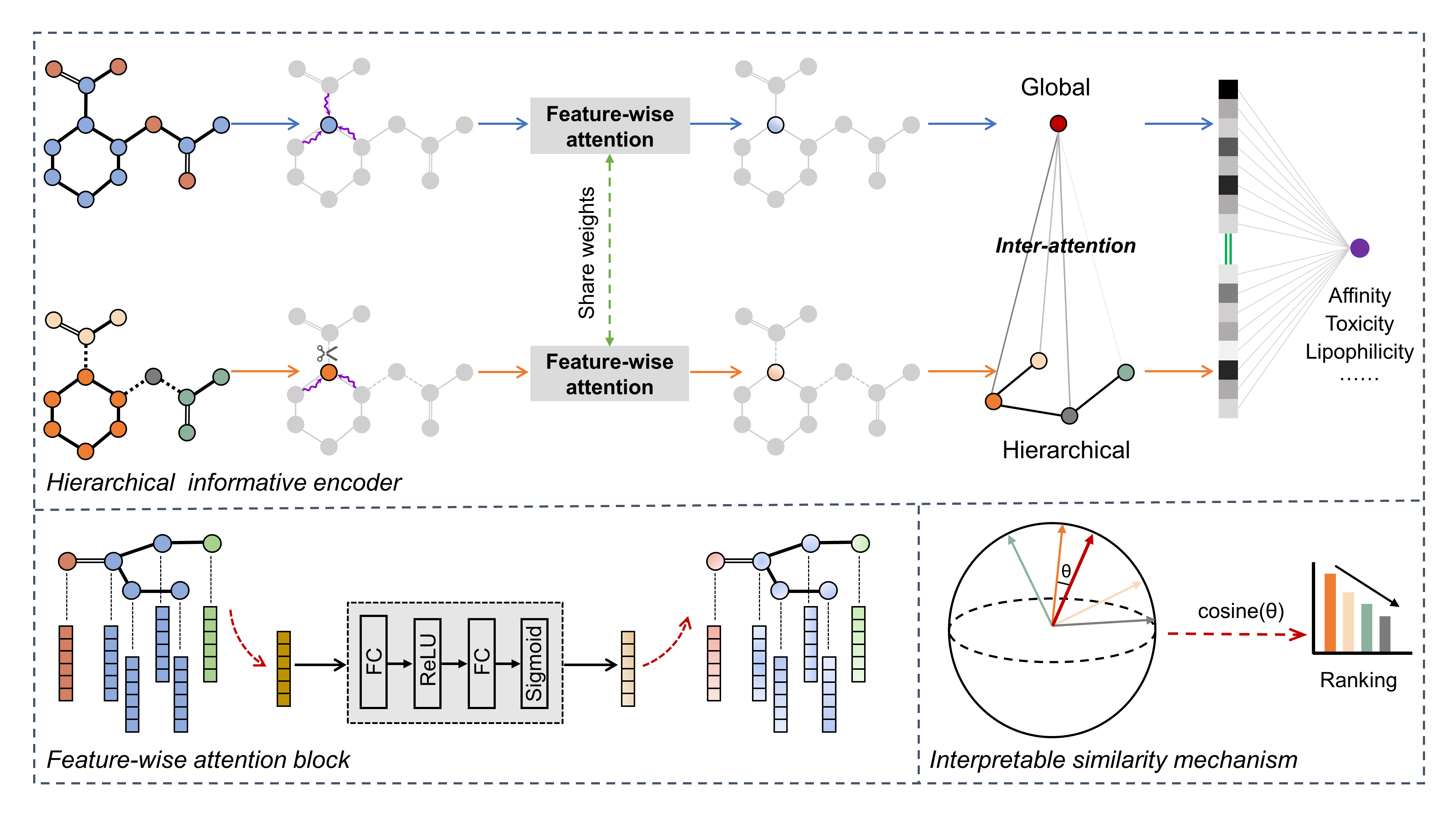
Elucidating and accurately predicting the druggability and bioactivities of molecules plays a pivotal role in drug design and discovery and remains an open challenge. Recently, graph neural networks (GNN) have made remarkable advancements in graph-based molecular property prediction. However, current graph-based deep learning methods neglect the hierarchical information of molecules and the relationships between feature channels. In this study, we propose a well-designed hierarchical informative graph neural networks framework (termed HiGNN) for predicting molecular property by utilizing a co-representation learning of molecular graphs and chemically synthesizable BRICS fragments. Furthermore, a plug-and-play feature-wise attention block is first designed in HiGNN architecture to adaptively recalibrate atomic features after the message passing phase. Extensive experiments demonstrate that HiGNN achieves state-of-the-art predictive performance on many challenging drug discovery-associated benchmark datasets. In addition, we devise a molecule-fragment similarity mechanism to comprehensively investigate the interpretability of HiGNN model at the subgraph level, indicating that HiGNN as a powerful deep learning tool can help chemists and pharmacists identify the key components of molecules for designing better molecules with desired properties or functions. The source code is publicly available at https://github.com/idruglab/hignn.
PDF Abstract