Interventions, Where and How? Experimental Design for Causal Models at Scale
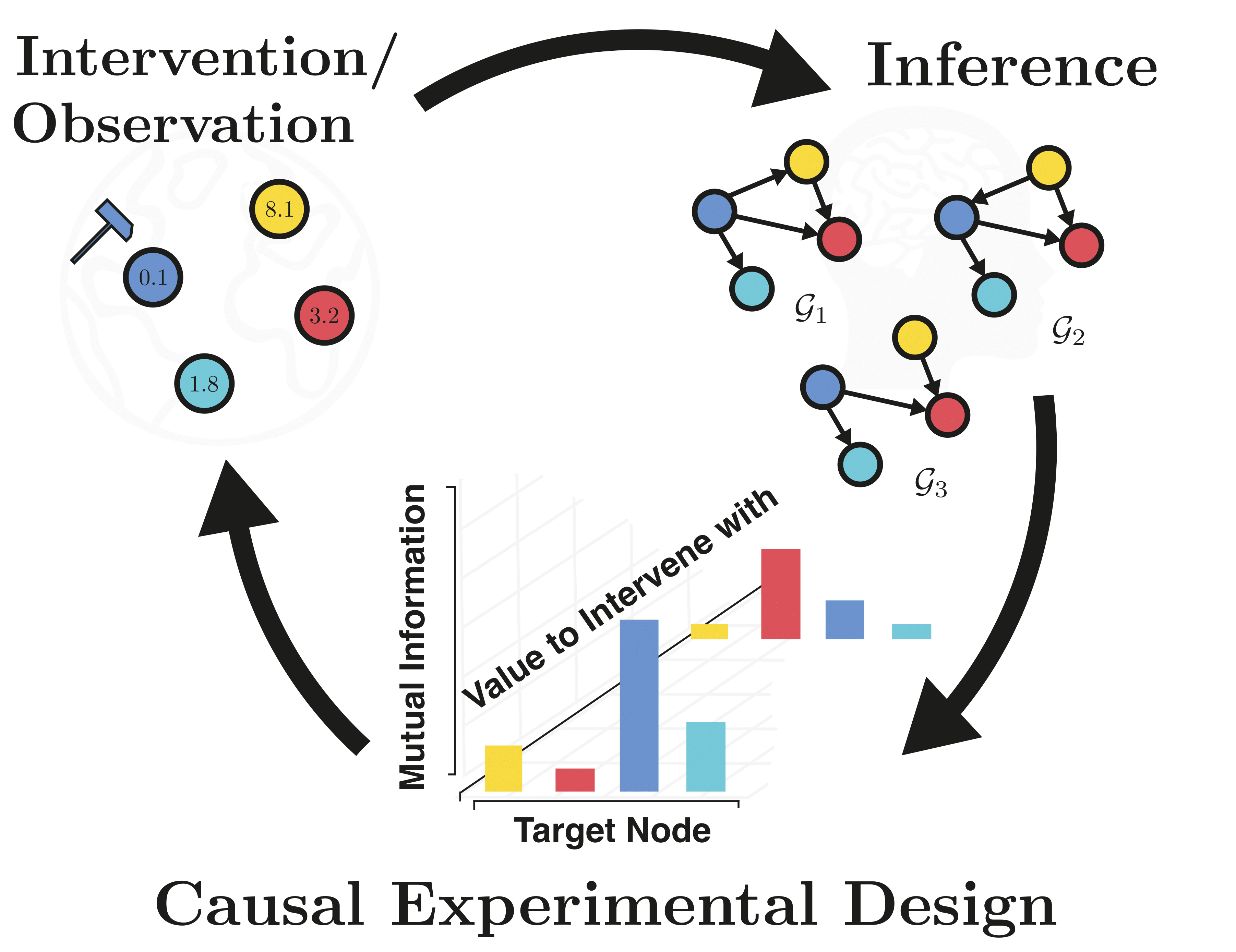
Causal discovery from observational and interventional data is challenging due to limited data and non-identifiability: factors that introduce uncertainty in estimating the underlying structural causal model (SCM). Selecting experiments (interventions) based on the uncertainty arising from both factors can expedite the identification of the SCM. Existing methods in experimental design for causal discovery from limited data either rely on linear assumptions for the SCM or select only the intervention target. This work incorporates recent advances in Bayesian causal discovery into the Bayesian optimal experimental design framework, allowing for active causal discovery of large, nonlinear SCMs while selecting both the interventional target and the value. We demonstrate the performance of the proposed method on synthetic graphs (Erdos-R\`enyi, Scale Free) for both linear and nonlinear SCMs as well as on the \emph{in-silico} single-cell gene regulatory network dataset, DREAM.
PDF Abstract