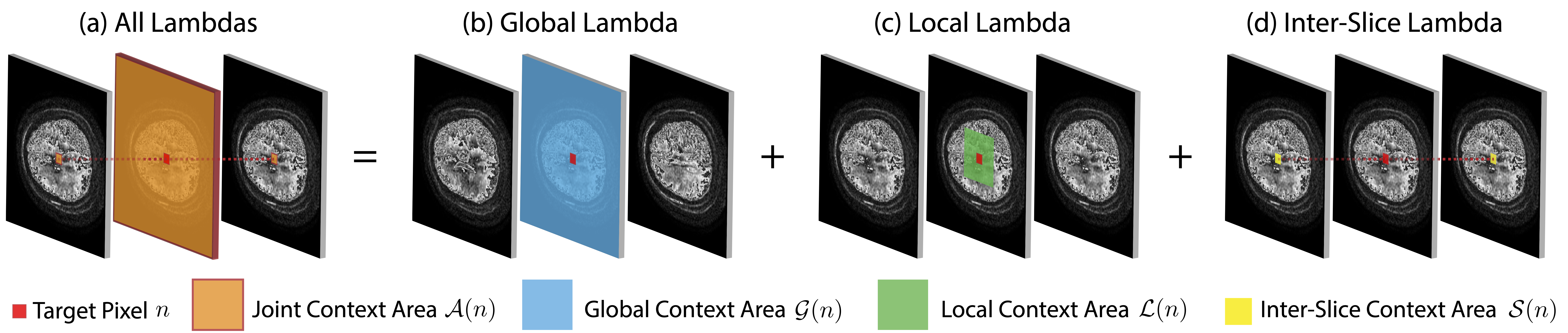
LambdaUNet: 2.5D Stroke Lesion Segmentation of Diffusion-weighted MR Images
Diffusion-weighted (DW) magnetic resonance imaging is essential for the diagnosis and treatment of ischemic stroke. DW images (DWIs) are usually acquired in multi-slice settings where lesion areas in two consecutive 2D slices are highly discontinuous due to large slice thickness and sometimes even slice gaps. Therefore, although DWIs contain rich 3D information, they cannot be treated as regular 3D or 2D images. Instead, DWIs are somewhere in-between (or 2.5D) due to the volumetric nature but inter-slice discontinuities. Thus, it is not ideal to apply most existing segmentation methods as they are designed for either 2D or 3D images. To tackle this problem, we propose a new neural network architecture tailored for segmenting highly-discontinuous 2.5D data such as DWIs. Our network, termed LambdaUNet, extends UNet by replacing convolutional layers with our proposed Lambda+ layers. In particular, Lambda+ layers transform both intra-slice and inter-slice context around a pixel into linear functions, called lambdas, which are then applied to the pixel to produce informative 2.5D features. LambdaUNet is simple yet effective in combining sparse inter-slice information from adjacent slices while also capturing dense contextual features within a single slice. Experiments on a unique clinical dataset demonstrate that LambdaUNet outperforms existing 3D/2D image segmentation methods including recent variants of UNet. Code for LambdaUNet is released with the publication to facilitate future research.
PDF Abstract