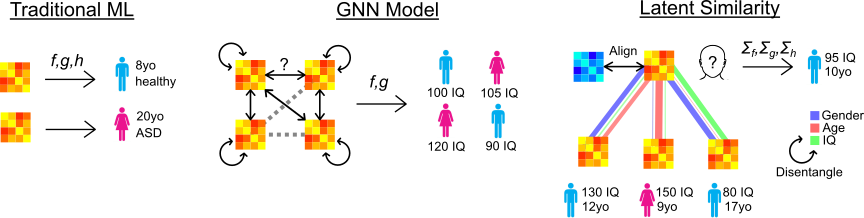Latent Similarity Identifies Important Functional Connections for Phenotype Prediction
Objective: Endophenotypes such as brain age and fluid intelligence are important biomarkers of disease status. However, brain imaging studies to identify these biomarkers often encounter limited numbers of subjects and high dimensional imaging features, hindering reproducibility. Therefore, we develop an interpretable, multivariate classification/regression algorithm, called Latent Similarity (LatSim), suitable for small sample size, high feature dimension datasets. Methods: LatSim combines metric learning with a kernel similarity function and softmax aggregation to identify task-related similarities between subjects. Inter-subject similarity is utilized to improve performance on three prediction tasks using multi-paradigm fMRI data. A greedy selection algorithm, made possible by LatSim's computational efficiency, is developed as an interpretability method. Results: LatSim achieved significantly higher predictive accuracy at small sample sizes on the Philadelphia Neurodevelopmental Cohort (PNC) dataset. Connections identified by LatSim gave superior discriminative power compared to those identified by other methods. We identified 4 functional brain networks enriched in connections for predicting brain age, sex, and intelligence. Conclusion: We find that most information for a predictive task comes from only a few (1-5) connections. Additionally, we find that the default mode network is over-represented in the top connections of all predictive tasks. Significance: We propose a novel algorithm for small sample, high feature dimension datasets and use it to identify connections in task fMRI data. Our work should lead to new insights in both algorithm design and neuroscience research. Code and demo are available at https://github.com/aorliche/LatentSimilarity/.
PDF Abstract