Map3D: Registration Based Multi-Object Tracking on 3D Serial Whole Slide Images
There has been a long pursuit for precise and reproducible glomerular quantification on renal pathology to leverage both research and practice. When digitizing the biopsy tissue samples using whole slide imaging (WSI), a set of serial sections from the same tissue can be acquired as a stack of images, similar to frames in a video. In radiology, the stack of images (e.g., computed tomography) are naturally used to provide 3D context for organs, tissues, and tumors. In pathology, it is appealing to do a similar 3D assessment. However, the 3D identification and association of large-scale glomeruli on renal pathology is challenging due to large tissue deformation, missing tissues, and artifacts from WSI. In this paper, we propose a novel Multi-object Association for Pathology in 3D (Map3D) method for automatically identifying and associating large-scale cross-sections of 3D objects from routine serial sectioning and WSI. The innovations of the Map3D method are three-fold: (1) the large-scale glomerular association is formed as a new multi-object tracking (MOT) perspective; (2) the quality-aware whole series registration is proposed to not only provide affinity estimation but also offer automatic kidney-wise quality assurance (QA) for registration; (3) a dual-path association method is proposed to tackle the large deformation, missing tissues, and artifacts during tracking. To the best of our knowledge, the Map3D method is the first approach that enables automatic and large-scale glomerular association across 3D serial sectioning using WSI. Our proposed method Map3D achieved MOTA= 44.6, which is 12.1% higher than the non deep learning benchmarks.
PDF Abstract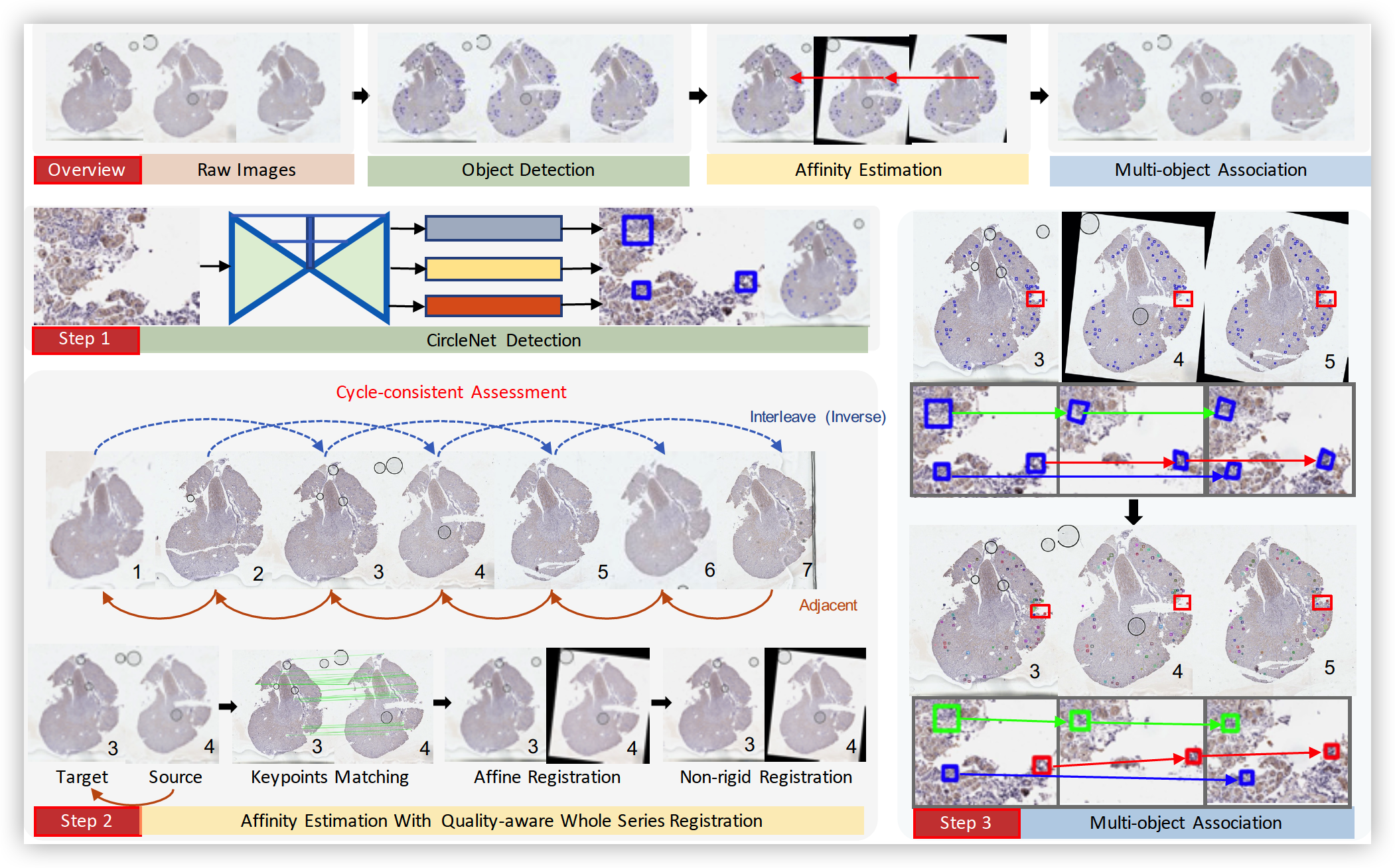



 MOTChallenge
MOTChallenge