MetaMedSeg: Volumetric Meta-learning for Few-Shot Organ Segmentation
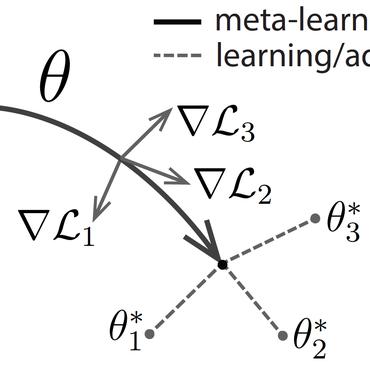
The lack of sufficient annotated image data is a common issue in medical image segmentation. For some organs and densities, the annotation may be scarce, leading to poor model training convergence, while other organs have plenty of annotated data. In this work, we present MetaMedSeg, a gradient-based meta-learning algorithm that redefines the meta-learning task for the volumetric medical data with the goal to capture the variety between the slices. We also explore different weighting schemes for gradients aggregation, arguing that different tasks might have different complexity, and hence, contribute differently to the initialization. We propose an importance-aware weighting scheme to train our model. In the experiments, we present an evaluation of the medical decathlon dataset by extracting 2D slices from CT and MRI volumes of different organs and performing semantic segmentation. The results show that our proposed volumetric task definition leads to up to 30% improvement in terms of IoU compared to related baselines. The proposed update rule is also shown to improve the performance for complex scenarios where the data distribution of the target organ is very different from the source organs.
PDF Abstract