Pre-training via Denoising for Molecular Property Prediction
Many important problems involving molecular property prediction from 3D structures have limited data, posing a generalization challenge for neural networks. In this paper, we describe a pre-training technique based on denoising that achieves a new state-of-the-art in molecular property prediction by utilizing large datasets of 3D molecular structures at equilibrium to learn meaningful representations for downstream tasks. Relying on the well-known link between denoising autoencoders and score-matching, we show that the denoising objective corresponds to learning a molecular force field -- arising from approximating the Boltzmann distribution with a mixture of Gaussians -- directly from equilibrium structures. Our experiments demonstrate that using this pre-training objective significantly improves performance on multiple benchmarks, achieving a new state-of-the-art on the majority of targets in the widely used QM9 dataset. Our analysis then provides practical insights into the effects of different factors -- dataset sizes, model size and architecture, and the choice of upstream and downstream datasets -- on pre-training.
PDF Abstract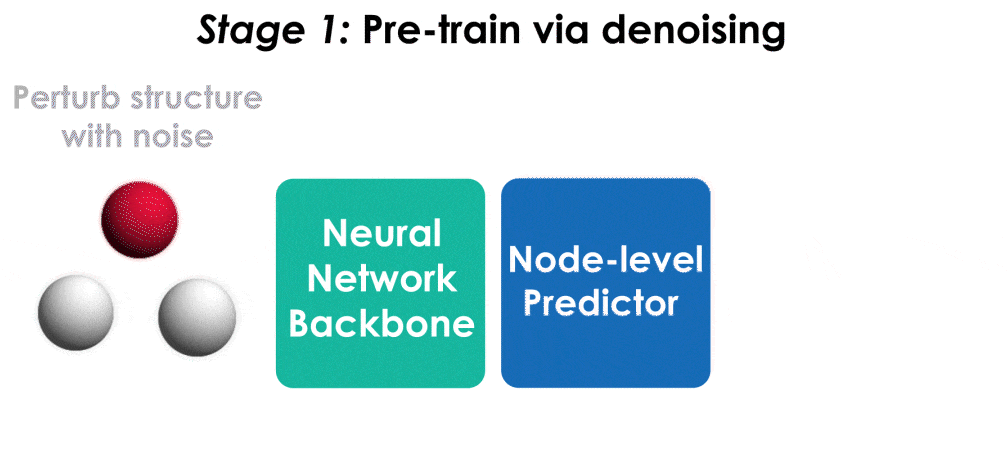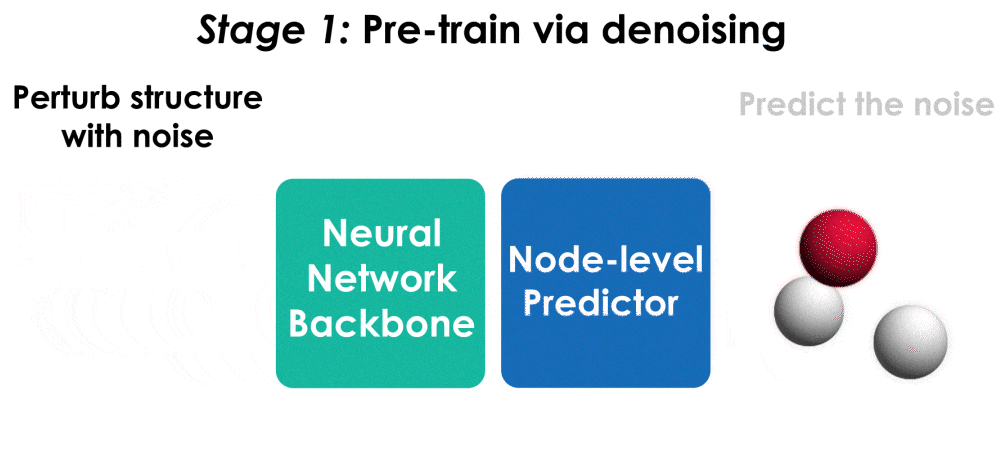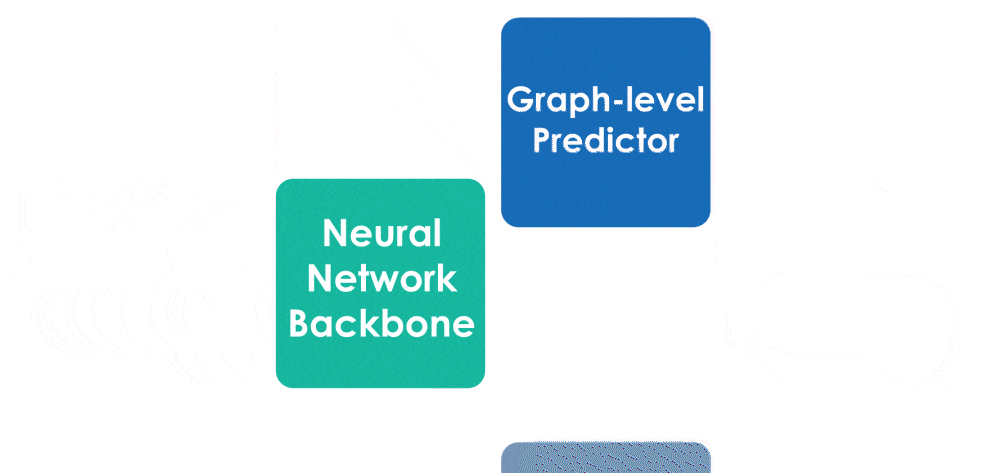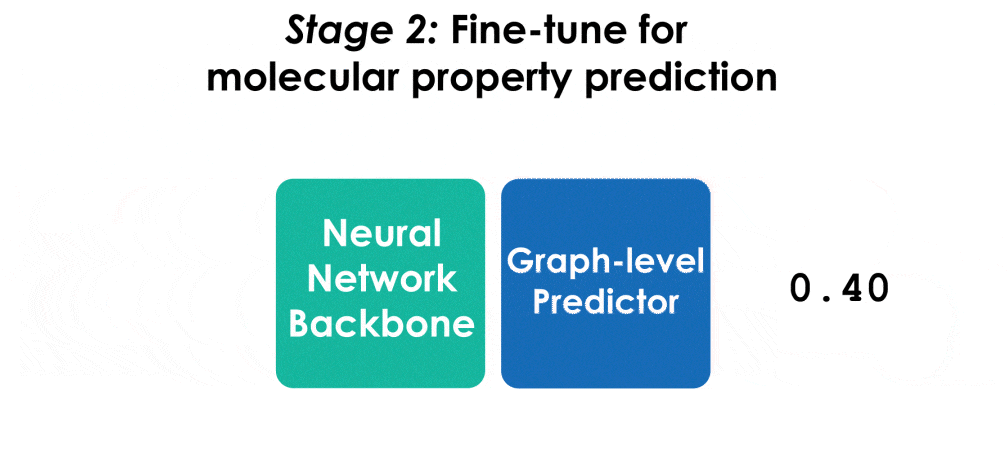



 OC20
OC20