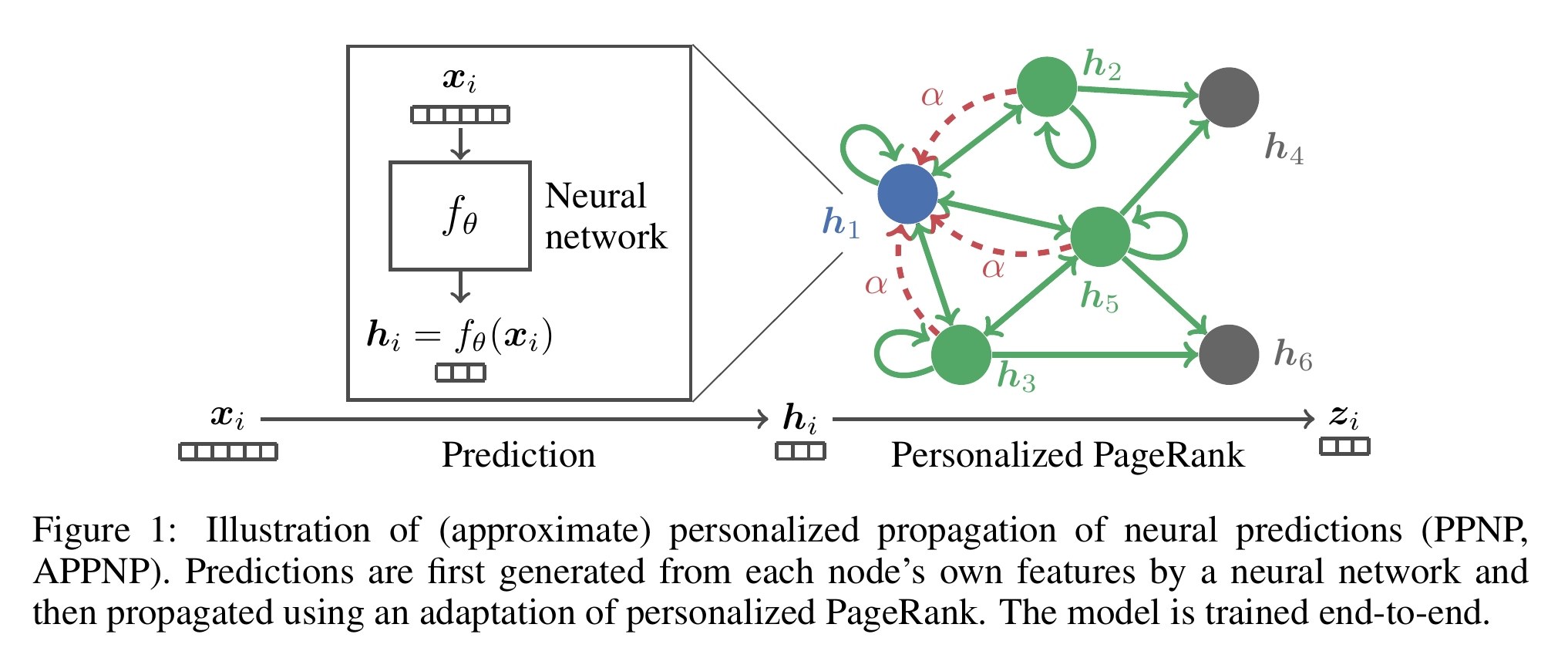
Predict then Propagate: Graph Neural Networks meet Personalized PageRank
Neural message passing algorithms for semi-supervised classification on graphs have recently achieved great success. However, for classifying a node these methods only consider nodes that are a few propagation steps away and the size of this utilized neighborhood is hard to extend. In this paper, we use the relationship between graph convolutional networks (GCN) and PageRank to derive an improved propagation scheme based on personalized PageRank. We utilize this propagation procedure to construct a simple model, personalized propagation of neural predictions (PPNP), and its fast approximation, APPNP. Our model's training time is on par or faster and its number of parameters on par or lower than previous models. It leverages a large, adjustable neighborhood for classification and can be easily combined with any neural network. We show that this model outperforms several recently proposed methods for semi-supervised classification in the most thorough study done so far for GCN-like models. Our implementation is available online.
PDF Abstract ICLR 2019 PDF ICLR 2019 AbstractCode
Datasets
 Cora
Citeseer
Penn94
genius
twitch-gamers
Deezer-Europe
Squirrel (60%/20%/20% random splits)
Wisconsin(60%/20%/20% random splits)
Cornell (60%/20%/20% random splits)
Texas(60%/20%/20% random splits)
PubMed (60%/20%/20% random splits)
Film (60%/20%/20% random splits)
Chameleon(60%/20%/20% random splits)
Cora
Citeseer
Penn94
genius
twitch-gamers
Deezer-Europe
Squirrel (60%/20%/20% random splits)
Wisconsin(60%/20%/20% random splits)
Cornell (60%/20%/20% random splits)
Texas(60%/20%/20% random splits)
PubMed (60%/20%/20% random splits)
Film (60%/20%/20% random splits)
Chameleon(60%/20%/20% random splits)