Protein Secondary Structure Prediction Using Cascaded Convolutional and Recurrent Neural Networks
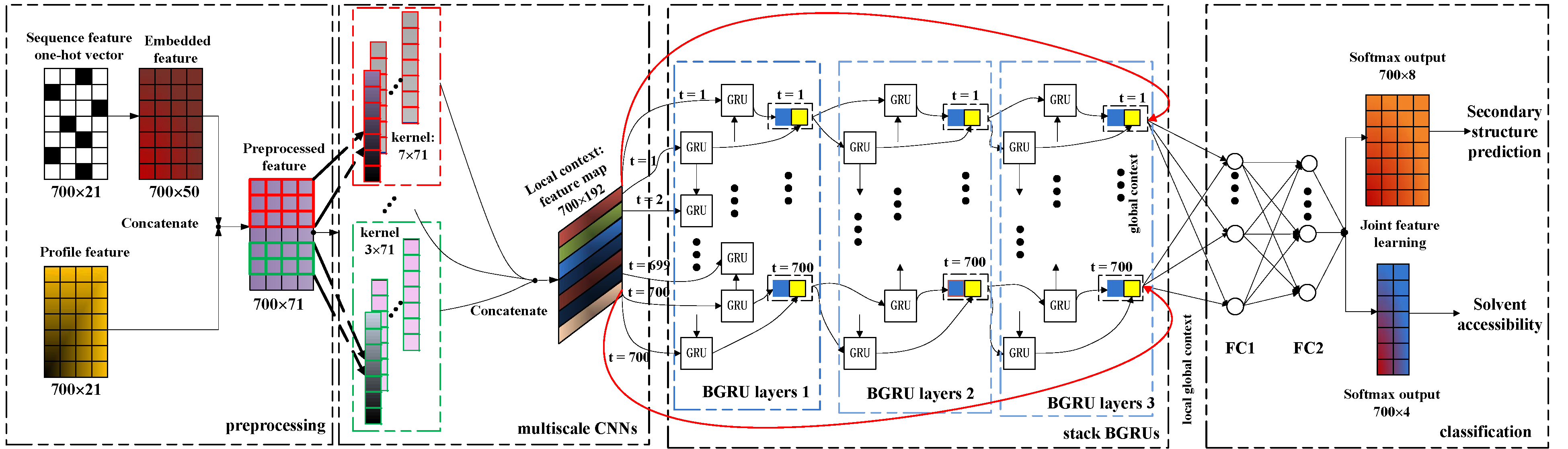
Protein secondary structure prediction is an important problem in bioinformatics. Inspired by the recent successes of deep neural networks, in this paper, we propose an end-to-end deep network that predicts protein secondary structures from integrated local and global contextual features. Our deep architecture leverages convolutional neural networks with different kernel sizes to extract multiscale local contextual features. In addition, considering long-range dependencies existing in amino acid sequences, we set up a bidirectional neural network consisting of gated recurrent unit to capture global contextual features. Furthermore, multi-task learning is utilized to predict secondary structure labels and amino-acid solvent accessibility simultaneously. Our proposed deep network demonstrates its effectiveness by achieving state-of-the-art performance, i.e., 69.7% Q8 accuracy on the public benchmark CB513, 76.9% Q8 accuracy on CASP10 and 73.1% Q8 accuracy on CASP11. Our model and results are publicly available.
PDF Abstract