Scalable learning for bridging the species gap in image-based plant phenotyping
The traditional paradigm of applying deep learning -- collect, annotate and train on data -- is not applicable to image-based plant phenotyping as almost 400,000 different plant species exists. Data costs include growing physical samples, imaging and labelling them. Model performance is impacted by the species gap between the domain of each plant species, it is not generalisable and may not transfer to unseen plant species. In this paper, we investigate the use of synthetic data for leaf instance segmentation. We study multiple synthetic data training regimes using Mask-RCNN when few or no annotated real data is available. We also present UPGen: a Universal Plant Generator for bridging the species gap. UPGen leverages domain randomisation to produce widely distributed data samples and models stochastic biological variation. Our methods outperform standard practices, such as transfer learning from publicly available plant data, by 26.6% and 51.46% on two unseen plant species respectively. We benchmark UPGen by competing in the CVPPP Leaf Segmentation Challenge and set a new state-of-the-art, a mean of 88% across A1-4 test datasets. This study is applicable to use of synthetic data for automating the measurement of phenotypic traits. Our synthetic dataset and pretrained model are available at https://csiro-robotics.github.io/UPGen_Webpage/.
PDF Abstract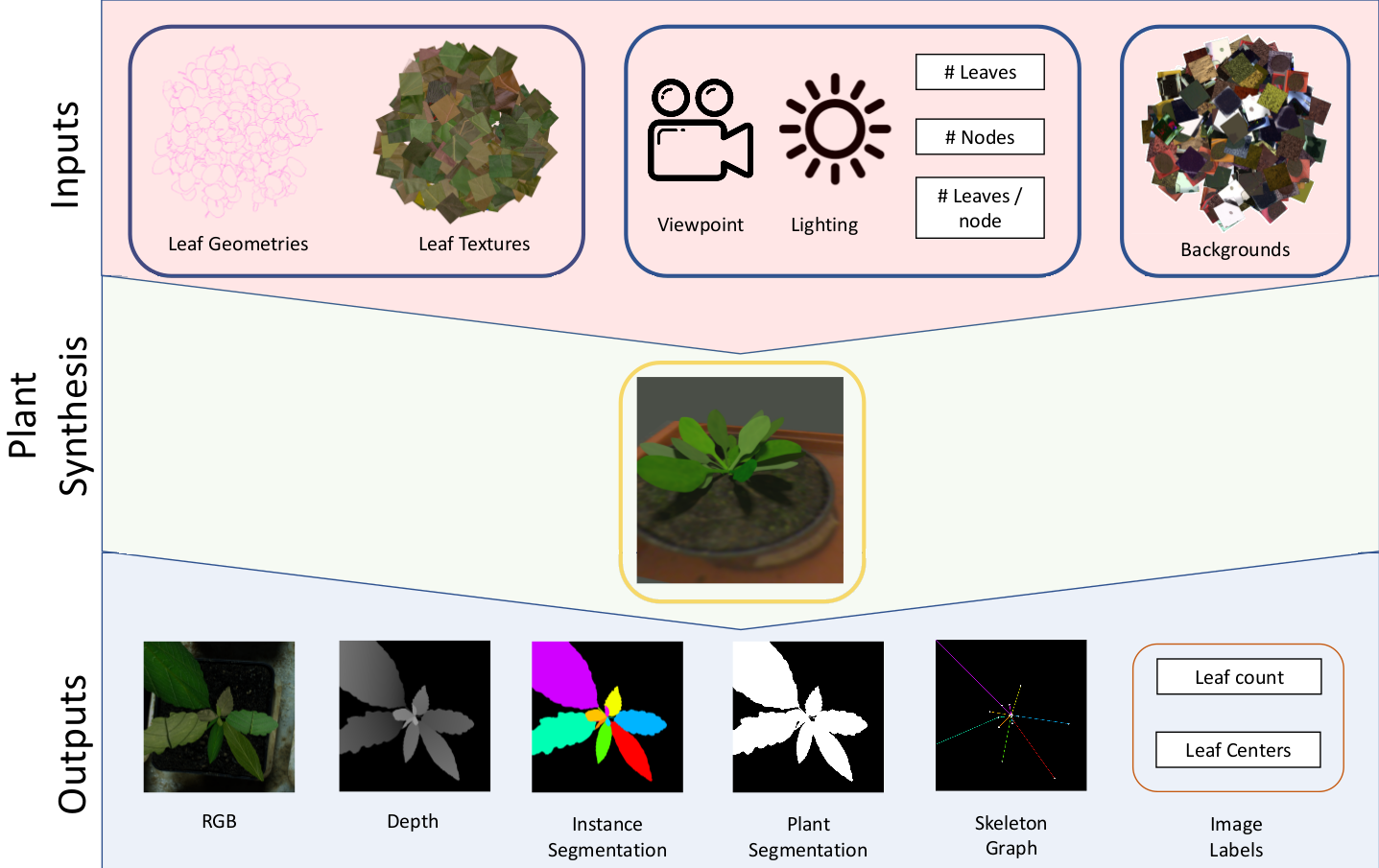




 MS COCO
MS COCO