Sequence-guided protein structure determination using graph convolutional and recurrent networks
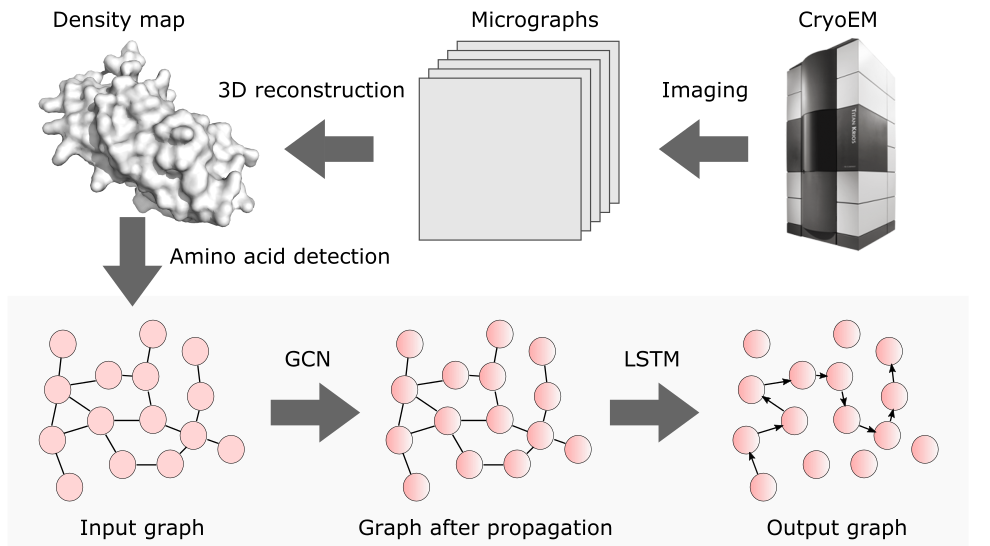
Single particle, cryogenic electron microscopy (cryo-EM) experiments now routinely produce high-resolution data for large proteins and their complexes. Building an atomic model into a cryo-EM density map is challenging, particularly when no structure for the target protein is known a priori. Existing protocols for this type of task often rely on significant human intervention and can take hours to many days to produce an output. Here, we present a fully automated, template-free model building approach that is based entirely on neural networks. We use a graph convolutional network (GCN) to generate an embedding from a set of rotamer-based amino acid identities and candidate 3-dimensional C$\alpha$ locations. Starting from this embedding, we use a bidirectional long short-term memory (LSTM) module to order and label the candidate identities and atomic locations consistent with the input protein sequence to obtain a structural model. Our approach paves the way for determining protein structures from cryo-EM densities at a fraction of the time of existing approaches and without the need for human intervention.
PDF Abstract