Shallow vs deep learning architectures for white matter lesion segmentation in the early stages of multiple sclerosis
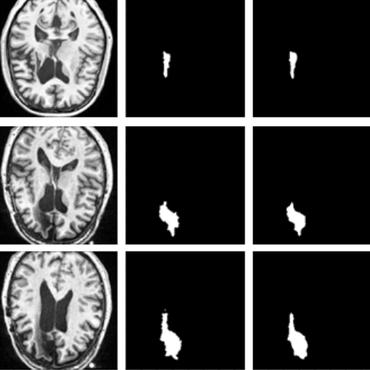
In this work, we present a comparison of a shallow and a deep learning architecture for the automated segmentation of white matter lesions in MR images of multiple sclerosis patients. In particular, we train and test both methods on early stage disease patients, to verify their performance in challenging conditions, more similar to a clinical setting than what is typically provided in multiple sclerosis segmentation challenges. Furthermore, we evaluate a prototype naive combination of the two methods, which refines the final segmentation. All methods were trained on 32 patients, and the evaluation was performed on a pure test set of 73 cases. Results show low lesion-wise false positives (30%) for the deep learning architecture, whereas the shallow architecture yields the best Dice coefficient (63%) and volume difference (19%). Combining both shallow and deep architectures further improves the lesion-wise metrics (69% and 26% lesion-wise true and false positive rate, respectively).
PDF Abstract