SleepEEGNet: Automated Sleep Stage Scoring with Sequence to Sequence Deep Learning Approach
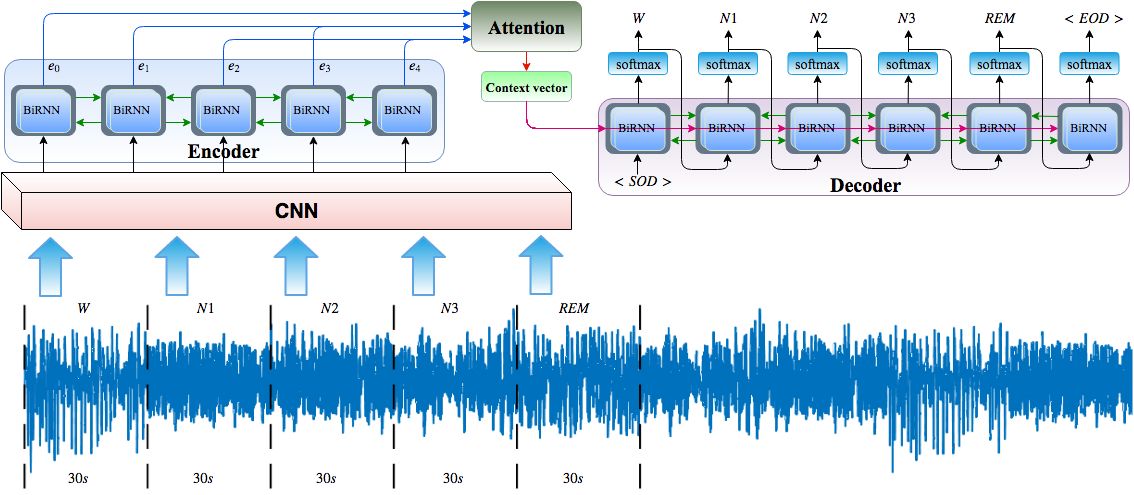
Electroencephalogram (EEG) is a common base signal used to monitor brain activity and diagnose sleep disorders. Manual sleep stage scoring is a time-consuming task for sleep experts and is limited by inter-rater reliability. In this paper, we propose an automatic sleep stage annotation method called SleepEEGNet using a single-channel EEG signal. The SleepEEGNet is composed of deep convolutional neural networks (CNNs) to extract time-invariant features, frequency information, and a sequence to sequence model to capture the complex and long short-term context dependencies between sleep epochs and scores. In addition, to reduce the effect of the class imbalance problem presented in the available sleep datasets, we applied novel loss functions to have an equal misclassified error for each sleep stage while training the network. We evaluated the proposed method on different single-EEG channels (i.e., Fpz-Cz and Pz-Oz EEG channels) from the Physionet Sleep-EDF datasets published in 2013 and 2018. The evaluation results demonstrate that the proposed method achieved the best annotation performance compared to current literature, with an overall accuracy of 84.26%, a macro F1-score of 79.66% and Cohen's Kappa coefficient = 0.79. Our developed model is ready to test with more sleep EEG signals and aid the sleep specialists to arrive at an accurate diagnosis. The source code is available at https://github.com/SajadMo/SleepEEGNet.
PDF Abstract