Small Lesion Segmentation in Brain MRIs with Subpixel Embedding
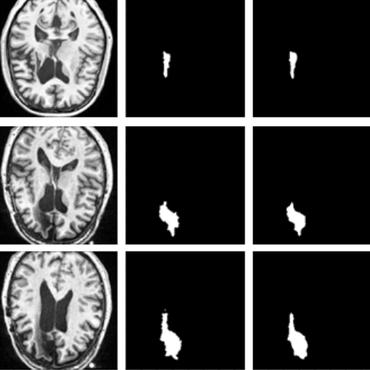
We present a method to segment MRI scans of the human brain into ischemic stroke lesion and normal tissues. We propose a neural network architecture in the form of a standard encoder-decoder where predictions are guided by a spatial expansion embedding network. Our embedding network learns features that can resolve detailed structures in the brain without the need for high-resolution training images, which are often unavailable and expensive to acquire. Alternatively, the encoder-decoder learns global structures by means of striding and max pooling. Our embedding network complements the encoder-decoder architecture by guiding the decoder with fine-grained details lost to spatial downsampling during the encoder stage. Unlike previous works, our decoder outputs at 2 times the input resolution, where a single pixel in the input resolution is predicted by four neighboring subpixels in our output. To obtain the output at the original scale, we propose a learnable downsampler (as opposed to hand-crafted ones e.g. bilinear) that combines subpixel predictions. Our approach improves the baseline architecture by approximately 11.7% and achieves the state of the art on the ATLAS public benchmark dataset with a smaller memory footprint and faster runtime than the best competing method. Our source code has been made available at: https://github.com/alexklwong/subpixel-embedding-segmentation.
PDF Abstract