Spectral Multigraph Networks for Discovering and Fusing Relationships in Molecules
Spectral Graph Convolutional Networks (GCNs) are a generalization of convolutional networks to learning on graph-structured data. Applications of spectral GCNs have been successful, but limited to a few problems where the graph is fixed, such as shape correspondence and node classification. In this work, we address this limitation by revisiting a particular family of spectral graph networks, Chebyshev GCNs, showing its efficacy in solving graph classification tasks with a variable graph structure and size. Chebyshev GCNs restrict graphs to have at most one edge between any pair of nodes. To this end, we propose a novel multigraph network that learns from multi-relational graphs. We model learned edges with abstract meaning and experiment with different ways to fuse the representations extracted from annotated and learned edges, achieving competitive results on a variety of chemical classification benchmarks.
PDF Abstract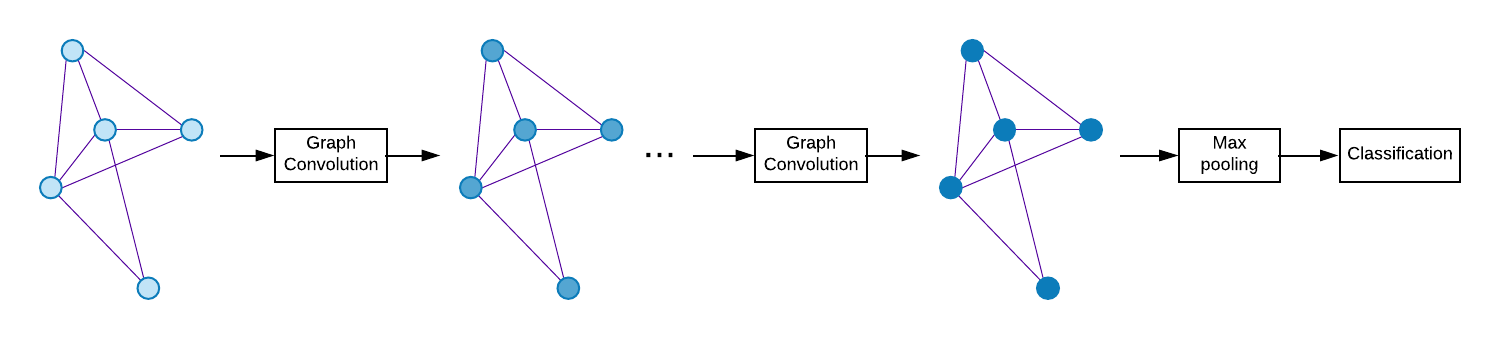










 NCI109
NCI109
