Systems Biology: Identifiability analysis and parameter identification via systems-biology informed neural networks
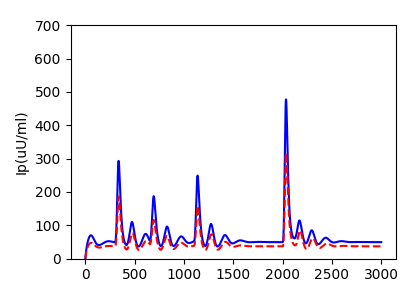
The dynamics of systems biological processes are usually modeled by a system of ordinary differential equations (ODEs) with many unknown parameters that need to be inferred from noisy and sparse measurements. Here, we introduce systems-biology informed neural networks for parameter estimation by incorporating the system of ODEs into the neural networks. To complete the workflow of system identification, we also describe structural and practical identifiability analysis to analyze the identifiability of parameters. We use the ultridian endocrine model for glucose-insulin interaction as the example to demonstrate all these methods and their implementation.
PDF Abstract