ULTRA: Uncertainty-aware Label Distribution Learning for Breast Tumor Cellularity Assessment
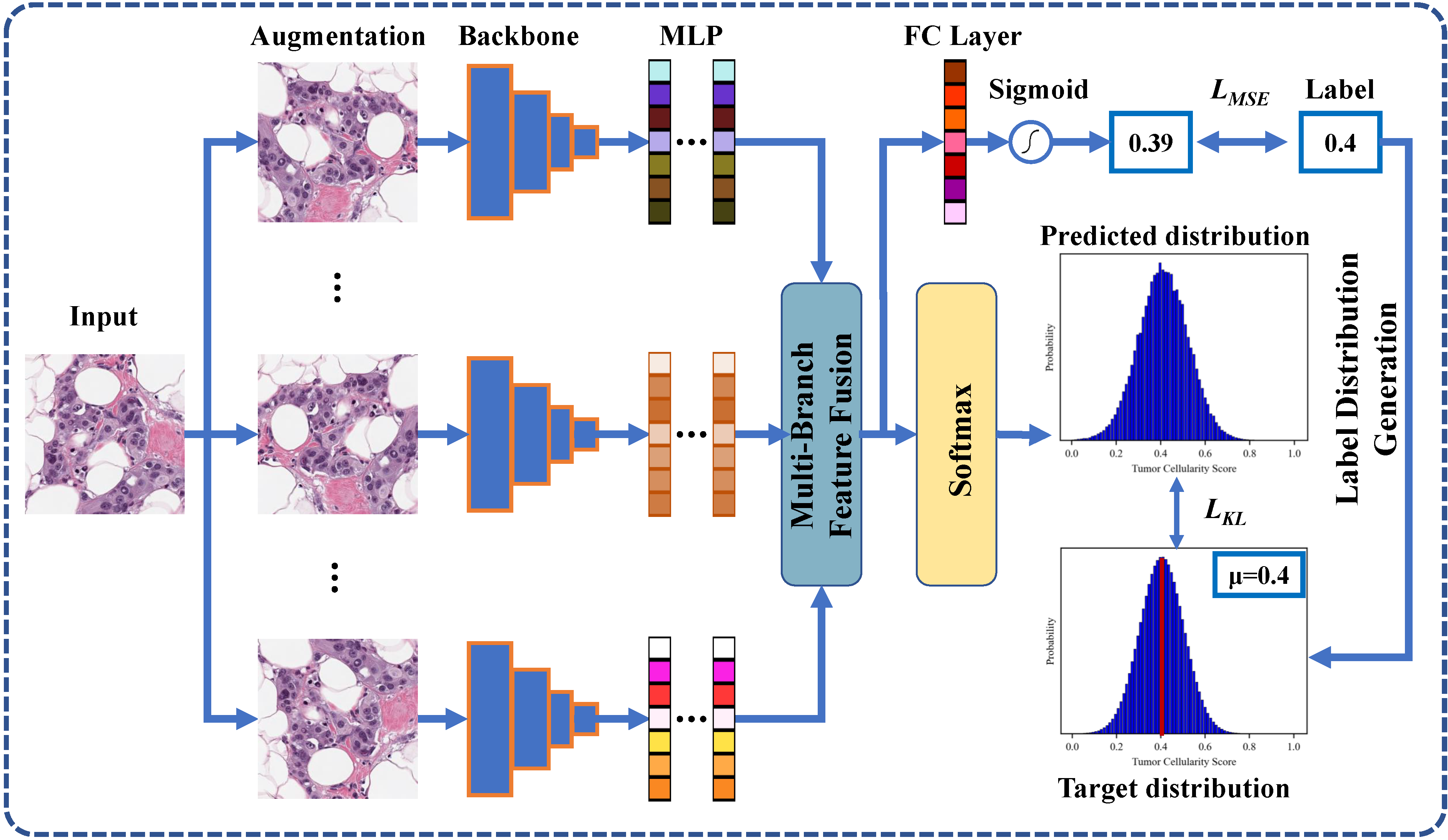
Neoadjuvant therapy (NAT) for breast cancer is a common treatment option in clinical practice. Tumor cellularity (TC), which represents the percentage of invasive tumors in the tumor bed, has been widely used to quantify the response of breast cancer to NAT. Therefore, automatic TC estimation is significant in clinical practice. However, existing state-of-the-art methods usually take it as a TC score regression problem, which ignores the ambiguity of TC labels caused by subjective assessment or multiple raters. In this paper, to efficiently leverage the label ambiguities, we proposed an Uncertainty-aware Label disTRibution leArning (ULTRA) framework for automatic TC estimation. The proposed ULTRA first converted the single-value TC labels to discrete label distributions, which effectively models the ambiguity among all possible TC labels. Furthermore, the network learned TC label distributions by minimizing the Kullback-Leibler (KL) divergence between the predicted and ground-truth TC label distributions, which better supervised the model to leverage the ambiguity of TC labels. Moreover, the ULTRA mimicked the multi-rater fusion process in clinical practice with a multi-branch feature fusion module to further explore the uncertainties of TC labels. We evaluated the ULTRA on the public BreastPathQ dataset. The experimental results demonstrate that the ULTRA outperformed the regression-based methods for a large margin and achieved state-of-the-art results. The code will be available from https://github.com/PerceptionComputingLab/ULTRA
PDF Abstract