Search Results for author: Faisal Mahmood
Found 39 papers, 20 papers with code
Graph Based Sinogram Denoising for Tomographic Reconstructions
no code implementations • 14 Mar 2016 • Faisal Mahmood, Nauman Shahid, Pierre Vandergheynst, Ulf Skoglund
This makes the sinogram an ideal candidate for graph based denoising since it generally has a piecewise smooth structure.
2D Discrete Fourier Transform with Simultaneous Edge Artifact Removal for Real-Time Applications
no code implementations • 16 Mar 2016 • Faisal Mahmood, Märt Toots, Lars-Göran Öfverstedt, Ulf Skoglund
These artifacts can have critical consequences if the DFTs are being used for further processing.
Adaptive Graph-based Total Variation for Tomographic Reconstructions
no code implementations • 4 Oct 2016 • Faisal Mahmood, Nauman Shahid, Ulf Skoglund, Pierre Vandergheynst
Sparsity exploiting image reconstruction (SER) methods have been extensively used with Total Variation (TV) regularization for tomographic reconstructions.
Deep Learning and Conditional Random Fields-based Depth Estimation and Topographical Reconstruction from Conventional Endoscopy
no code implementations • 30 Oct 2017 • Faisal Mahmood, Nicholas J. Durr
We show that the estimated depth maps can be used for reconstructing the topography of the mucosa from conventional colonoscopy images.
Unsupervised Reverse Domain Adaptation for Synthetic Medical Images via Adversarial Training
no code implementations • 17 Nov 2017 • Faisal Mahmood, Richard Chen, Nicholas J. Durr
We propose an alternative framework that uses a reverse flow, where adversarial training is used to make real medical images more like synthetic images, and hypothesize that clinically-relevant features can be preserved via self-regularization.
Deep Learning with Cinematic Rendering: Fine-Tuning Deep Neural Networks Using Photorealistic Medical Images
no code implementations • 22 May 2018 • Faisal Mahmood, Richard Chen, Sandra Sudarsky, Daphne Yu, Nicholas J. Durr
Our experiments demonstrate that: (a) Convolutional Neural Networks (CNNs) trained on synthetic data and fine-tuned on photorealistic cinematically rendered data adapt better to real medical images and demonstrate more robust performance when compared to networks with no fine-tuning, (b) these fine-tuned networks require less training data to converge to an optimal solution, and (c) fine-tuning with data from a variety of photorealistic rendering conditions of the same scene prevents the network from learning patient-specific information and aids in generalizability of the model.
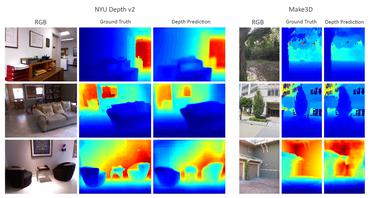
Rethinking Monocular Depth Estimation with Adversarial Training
no code implementations • 22 Aug 2018 • Richard Chen, Faisal Mahmood, Alan Yuille, Nicholas J. Durr
Most existing approaches treat depth estimation as a regression problem with a local pixel-wise loss function.
Deep Adversarial Training for Multi-Organ Nuclei Segmentation in Histopathology Images
1 code implementation • 29 Sep 2018 • Faisal Mahmood, Daniel Borders, Richard Chen, Gregory N. McKay, Kevan J. Salimian, Alexander Baras, Nicholas J. Durr
However, CNNs require large amounts of labeled histopathology data.
DeepLSR: a deep learning approach for laser speckle reduction
2 code implementations • 23 Oct 2018 • Taylor L. Bobrow, Faisal Mahmood, Miguel Inserni, Nicholas J. Durr
In images of gastrointestinal tissues, DeepLSR reduces laser speckle noise by 6. 4 dB, compared to a 2. 9 dB reduction from optimized non-local means processing, a 3. 0 dB reduction from BM3D, and a 3. 7 dB reduction from an optical speckle reducer utilizing an oscillating diffuser.
Multimodal Densenet
no code implementations • 18 Nov 2018 • Faisal Mahmood, Ziyun Yang, Thomas Ashley, Nicholas J. Durr
In this work, we propose Multimodal DenseNet, a novel architecture for fusing multimodal data.
Structured Prediction using cGANs with Fusion Discriminator
no code implementations • ICLR 2019 • Faisal Mahmood, Wenhao Xu, Nicholas J. Durr, Jeremiah W. Johnson, Alan Yuille
We propose the fusion discriminator, a single unified framework for incorporating conditional information into a generative adversarial network (GAN) for a variety of distinct structured prediction tasks, including image synthesis, semantic segmentation, and depth estimation.
GANPOP: Generative Adversarial Network Prediction of Optical Properties from Single Snapshot Wide-field Images
no code implementations • 12 Jun 2019 • Mason T. Chen, Faisal Mahmood, Jordan A. Sweer, Nicholas J. Durr
In human gastrointestinal specimens, GANPOP estimates both reduced scattering and absorption coefficients at 660 nm from a single 0. 2/mm spatial frequency illumination image with 58% higher accuracy than SSOP.
SLAM Endoscopy enhanced by adversarial depth prediction
no code implementations • 29 Jun 2019 • Richard J. Chen, Taylor L. Bobrow, Thomas Athey, Faisal Mahmood, Nicholas J. Durr
Medical endoscopy remains a challenging application for simultaneous localization and mapping (SLAM) due to the sparsity of image features and size constraints that prevent direct depth-sensing.
Semi-Supervised Histology Classification using Deep Multiple Instance Learning and Contrastive Predictive Coding
no code implementations • 23 Oct 2019 • Ming Y. Lu, Richard J. Chen, Jingwen Wang, Debora Dillon, Faisal Mahmood
Convolutional neural networks can be trained to perform histology slide classification using weak annotations with multiple instance learning (MIL).
Weakly Supervised Prostate TMA Classification via Graph Convolutional Networks
no code implementations • 29 Oct 2019 • Jingwen Wang, Richard J. Chen, Ming Y. Lu, Alexander Baras, Faisal Mahmood
In prostate cancer, the Gleason score is a grading system used to measure the aggressiveness of prostate cancer from the spatial organization of cells and the distribution of glands.
Pathomic Fusion: An Integrated Framework for Fusing Histopathology and Genomic Features for Cancer Diagnosis and Prognosis
1 code implementation • 18 Dec 2019 • Richard J. Chen, Ming Y. Lu, Jingwen Wang, Drew F. K. Williamson, Scott J. Rodig, Neal I. Lindeman, Faisal Mahmood
Cancer diagnosis, prognosis, and therapeutic response predictions are based on morphological information from histology slides and molecular profiles from genomic data.
EndoL2H: Deep Super-Resolution for Capsule Endoscopy
3 code implementations • 13 Feb 2020 • Yasin Almalioglu, Kutsev Bengisu Ozyoruk, Abdulkadir Gokce, Kagan Incetan, Guliz Irem Gokceler, Muhammed Ali Simsek, Kivanc Ararat, Richard J. Chen, Nicholas J. Durr, Faisal Mahmood, Mehmet Turan
Although wireless capsule endoscopy is the preferred modality for diagnosis and assessment of small bowel diseases, the poor camera resolution is a substantial limitation for both subjective and automated diagnostics.
Data Efficient and Weakly Supervised Computational Pathology on Whole Slide Images
1 code implementation • 20 Apr 2020 • Ming Y. Lu, Drew F. K. Williamson, Tiffany Y. Chen, Richard J. Chen, Matteo Barbieri, Faisal Mahmood
CLAM is a general-purpose and adaptable method that can be used for a variety of different computational pathology tasks in both clinical and research settings.
Deep Learning-based Computational Pathology Predicts Origins for Cancers of Unknown Primary
1 code implementation • 24 Jun 2020 • Ming Y. Lu, Melissa Zhao, Maha Shady, Jana Lipkova, Tiffany Y. Chen, Drew F. K. Williamson, Faisal Mahmood
Cancer of unknown primary (CUP) is an enigmatic group of diagnoses where the primary anatomical site of tumor origin cannot be determined.
EndoSLAM Dataset and An Unsupervised Monocular Visual Odometry and Depth Estimation Approach for Endoscopic Videos: Endo-SfMLearner
1 code implementation • 30 Jun 2020 • Kutsev Bengisu Ozyoruk, Guliz Irem Gokceler, Gulfize Coskun, Kagan Incetan, Yasin Almalioglu, Faisal Mahmood, Eva Curto, Luis Perdigoto, Marina Oliveira, Hasan Sahin, Helder Araujo, Henrique Alexandrino, Nicholas J. Durr, Hunter B. Gilbert, Mehmet Turan
The codes and the link for the dataset are publicly available at https://github. com/CapsuleEndoscope/EndoSLAM.
VR-Caps: A Virtual Environment for Capsule Endoscopy
2 code implementations • 29 Aug 2020 • Kagan Incetan, Ibrahim Omer Celik, Abdulhamid Obeid, Guliz Irem Gokceler, Kutsev Bengisu Ozyoruk, Yasin Almalioglu, Richard J. Chen, Faisal Mahmood, Hunter Gilbert, Nicholas J. Durr, Mehmet Turan
Current capsule endoscopes and next-generation robotic capsules for diagnosis and treatment of gastrointestinal diseases are complex cyber-physical platforms that must orchestrate complex software and hardware functions.
Federated Learning for Computational Pathology on Gigapixel Whole Slide Images
1 code implementation • 21 Sep 2020 • Ming Y. Lu, Dehan Kong, Jana Lipkova, Richard J. Chen, Rajendra Singh, Drew F. K. Williamson, Tiffany Y. Chen, Faisal Mahmood
In this paper, we introduce privacy-preserving federated learning for gigapixel whole slide images in computational pathology using weakly-supervised attention multiple instance learning and differential privacy.
Multimodal Co-Attention Transformer for Survival Prediction in Gigapixel Whole Slide Images
1 code implementation • ICCV 2021 • Richard J. Chen, Ming Y. Lu, Wei-Hung Weng, Tiffany Y. Chen, Drew F.K. Williamson, Trevor Manz, Maha Shady, Faisal Mahmood
Survival outcome prediction is a challenging weakly-supervised and ordinal regression task in computational pathology that involves modeling complex interactions within the tumor microenvironment in gigapixel whole slide images (WSIs).
Deep Learning-based Frozen Section to FFPE Translation
1 code implementation • 25 Jul 2021 • Kutsev Bengisu Ozyoruk, Sermet Can, Guliz Irem Gokceler, Kayhan Basak, Derya Demir, Gurdeniz Serin, Uguray Payam Hacisalihoglu, Emirhan Kurtuluş, Berkan Darbaz, Ming Y. Lu, Tiffany Y. Chen, Drew F. K. Williamson, Funda Yilmaz, Faisal Mahmood, Mehmet Turan
In this paper, we propose an artificial intelligence (AI) method that improves FS image quality by computationally transforming frozen-sectioned whole-slide images (FS-WSIs) into whole-slide FFPE-style images in minutes.
Whole Slide Images are 2D Point Clouds: Context-Aware Survival Prediction using Patch-based Graph Convolutional Networks
1 code implementation • 27 Jul 2021 • Richard J. Chen, Ming Y. Lu, Muhammad Shaban, Chengkuan Chen, Tiffany Y. Chen, Drew F. K. Williamson, Faisal Mahmood
Cancer prognostication is a challenging task in computational pathology that requires context-aware representations of histology features to adequately infer patient survival.
Fast and Scalable Image Search For Histology
2 code implementations • 28 Jul 2021 • Chengkuan Chen, Ming Y. Lu, Drew F. K. Williamson, Tiffany Y. Chen, Andrew J. Schaumberg, Faisal Mahmood
Similar pathology image search offers the opportunity to comb through large historical repositories of gigapixel WSIs to identify cases with similar morphological features and can be particularly useful for diagnosing rare diseases, identifying similar cases for predicting prognosis, treatment outcomes, and potential clinical trial success.
Pan-Cancer Integrative Histology-Genomic Analysis via Interpretable Multimodal Deep Learning
1 code implementation • 4 Aug 2021 • Richard J. Chen, Ming Y. Lu, Drew F. K. Williamson, Tiffany Y. Chen, Jana Lipkova, Muhammad Shaban, Maha Shady, Mane Williams, Bumjin Joo, Zahra Noor, Faisal Mahmood
To validate that these model explanations are prognostic, we further analyzed high attention morphological regions in WSIs, which indicates that tumor-infiltrating lymphocyte presence corroborates with favorable cancer prognosis on 9 out of 14 cancer types studied.
Algorithm Fairness in AI for Medicine and Healthcare
no code implementations • 1 Oct 2021 • Richard J. Chen, Tiffany Y. Chen, Jana Lipkova, Judy J. Wang, Drew F. K. Williamson, Ming Y. Lu, Sharifa Sahai, Faisal Mahmood
In the current development and deployment of many artificial intelligence (AI) systems in healthcare, algorithm fairness is a challenging problem in delivering equitable care.
Scaling Vision Transformers to Gigapixel Images via Hierarchical Self-Supervised Learning
2 code implementations • CVPR 2022 • Richard J. Chen, Chengkuan Chen, Yicong Li, Tiffany Y. Chen, Andrew D. Trister, Rahul G. Krishnan, Faisal Mahmood
Vision Transformers (ViTs) and their multi-scale and hierarchical variations have been successful at capturing image representations but their use has been generally studied for low-resolution images (e. g. - 256x256, 384384).
Incorporating intratumoral heterogeneity into weakly-supervised deep learning models via variance pooling
1 code implementation • 17 Jun 2022 • Iain Carmichael, Andrew H. Song, Richard J. Chen, Drew F. K. Williamson, Tiffany Y. Chen, Faisal Mahmood
Supervised learning tasks such as cancer survival prediction from gigapixel whole slide images (WSIs) are a critical challenge in computational pathology that requires modeling complex features of the tumor microenvironment.
Embedding Space Augmentation for Weakly Supervised Learning in Whole-Slide Images
no code implementations • 31 Oct 2022 • Imaad Zaffar, Guillaume Jaume, Nasir Rajpoot, Faisal Mahmood
Multiple Instance Learning (MIL) is a widely employed framework for learning on gigapixel whole-slide images (WSIs) from WSI-level annotations.
Quantifying & Modeling Multimodal Interactions: An Information Decomposition Framework
1 code implementation • NeurIPS 2023 • Paul Pu Liang, Yun Cheng, Xiang Fan, Chun Kai Ling, Suzanne Nie, Richard Chen, Zihao Deng, Nicholas Allen, Randy Auerbach, Faisal Mahmood, Ruslan Salakhutdinov, Louis-Philippe Morency
The recent explosion of interest in multimodal applications has resulted in a wide selection of datasets and methods for representing and integrating information from different modalities.
Modeling Dense Multimodal Interactions Between Biological Pathways and Histology for Survival Prediction
2 code implementations • 13 Apr 2023 • Guillaume Jaume, Anurag Vaidya, Richard Chen, Drew Williamson, Paul Liang, Faisal Mahmood
We propose fusing both modalities using a memory-efficient multimodal Transformer that can model interactions between pathway and histology patch tokens.
Visual Language Pretrained Multiple Instance Zero-Shot Transfer for Histopathology Images
1 code implementation • CVPR 2023 • Ming Y. Lu, Bowen Chen, Andrew Zhang, Drew F. K. Williamson, Richard J. Chen, Tong Ding, Long Phi Le, Yung-Sung Chuang, Faisal Mahmood
In this paper we present MI-Zero, a simple and intuitive framework for unleashing the zero-shot transfer capabilities of contrastively aligned image and text models on gigapixel histopathology whole slide images, enabling multiple downstream diagnostic tasks to be carried out by pretrained encoders without requiring any additional labels.
Towards a Visual-Language Foundation Model for Computational Pathology
no code implementations • 24 Jul 2023 • Ming Y. Lu, Bowen Chen, Drew F. K. Williamson, Richard J. Chen, Ivy Liang, Tong Ding, Guillaume Jaume, Igor Odintsov, Andrew Zhang, Long Phi Le, Georg Gerber, Anil V Parwani, Faisal Mahmood
The accelerated adoption of digital pathology and advances in deep learning have enabled the development of powerful models for various pathology tasks across a diverse array of diseases and patient cohorts.
Weakly Supervised AI for Efficient Analysis of 3D Pathology Samples
1 code implementation • 27 Jul 2023 • Andrew H. Song, Mane Williams, Drew F. K. Williamson, Guillaume Jaume, Andrew Zhang, Bowen Chen, Robert Serafin, Jonathan T. C. Liu, Alex Baras, Anil V. Parwani, Faisal Mahmood
Human tissue and its constituent cells form a microenvironment that is fundamentally three-dimensional (3D).
A General-Purpose Self-Supervised Model for Computational Pathology
no code implementations • 29 Aug 2023 • Richard J. Chen, Tong Ding, Ming Y. Lu, Drew F. K. Williamson, Guillaume Jaume, Bowen Chen, Andrew Zhang, Daniel Shao, Andrew H. Song, Muhammad Shaban, Mane Williams, Anurag Vaidya, Sharifa Sahai, Lukas Oldenburg, Luca L. Weishaupt, Judy J. Wang, Walt Williams, Long Phi Le, Georg Gerber, Faisal Mahmood
Tissue phenotyping is a fundamental computational pathology (CPath) task in learning objective characterizations of histopathologic biomarkers in anatomic pathology.
A Foundational Multimodal Vision Language AI Assistant for Human Pathology
no code implementations • 13 Dec 2023 • Ming Y. Lu, Bowen Chen, Drew F. K. Williamson, Richard J. Chen, Kenji Ikamura, Georg Gerber, Ivy Liang, Long Phi Le, Tong Ding, Anil V Parwani, Faisal Mahmood
We compare PathChat against several multimodal vision language AI assistants as well as GPT4V, which powers the commercially available multimodal general purpose AI assistant ChatGPT-4.
Artificial Intelligence for Digital and Computational Pathology
no code implementations • 13 Dec 2023 • Andrew H. Song, Guillaume Jaume, Drew F. K. Williamson, Ming Y. Lu, Anurag Vaidya, Tiffany R. Miller, Faisal Mahmood
Advances in digitizing tissue slides and the fast-paced progress in artificial intelligence, including deep learning, have boosted the field of computational pathology.