Search Results for author: Riccardo de Feo
Found 5 papers, 2 papers with code
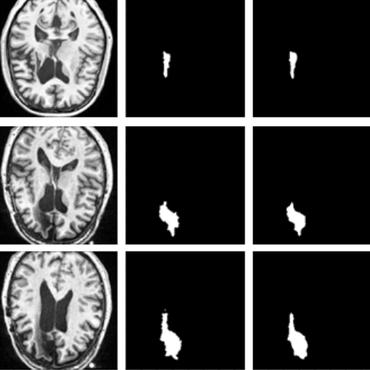
RatLesNetv2: A Fully Convolutional Network for Rodent Brain Lesion Segmentation
1 code implementation • 24 Jan 2020 • Juan Miguel Valverde, Artem Shatillo, Riccardo de Feo, Olli Gröhn, Alejandra Sierra, Jussi Tohka
RatLesNetv2 obtained similar to higher Dice coefficient values than the other ConvNets and it produced much more realistic and compact segmentations with notably fewer holes and lower Hausdorff distance.
Automatic cerebral hemisphere segmentation in rat MRI with lesions via attention-based convolutional neural networks
1 code implementation • 4 Aug 2021 • Juan Miguel Valverde, Artem Shatillo, Riccardo de Feo, Jussi Tohka
We present MedicDeepLabv3+, a convolutional neural network that is the first completely automatic method to segment cerebral hemispheres in magnetic resonance (MR) volumes of rats with lesions.
Automatic Rodent Brain MRI Lesion Segmentation with Fully Convolutional Networks
no code implementations • 23 Aug 2019 • Juan Miguel Valverde, Artem Shatillo, Riccardo de Feo, Olli Gröhn, Alejandra Sierra, Jussi Tohka
Several automatic methods have been developed for different human brain MRI segmentation, but little research has targeted automatic rodent lesion segmentation.
Transfer Learning in Magnetic Resonance Brain Imaging: a Systematic Review
no code implementations • 2 Feb 2021 • Juan Miguel Valverde, Vandad Imani, Ali Abdollahzadeh, Riccardo de Feo, Mithilesh Prakash, Robert Ciszek, Jussi Tohka
We found 129 articles that applied transfer learning to brain MRI tasks.
Convolutional Neural Networks for Automatic Detection of Intact Adenovirus from TEM Imaging with Debris, Broken and Artefacts Particles
no code implementations • 30 Oct 2023 • Olivier Rukundo, Andrea Behanova, Riccardo de Feo, Seppo Ronkko, Joni Oja, Jussi Tohka
To overcome the challenge, due to such a presence, we developed a software tool for semi-automatic annotation and segmentation of adenoviruses and a software tool for automatic segmentation and detection of intact adenoviruses in TEM imaging systems.