ChiENN: Embracing Molecular Chirality with Graph Neural Networks
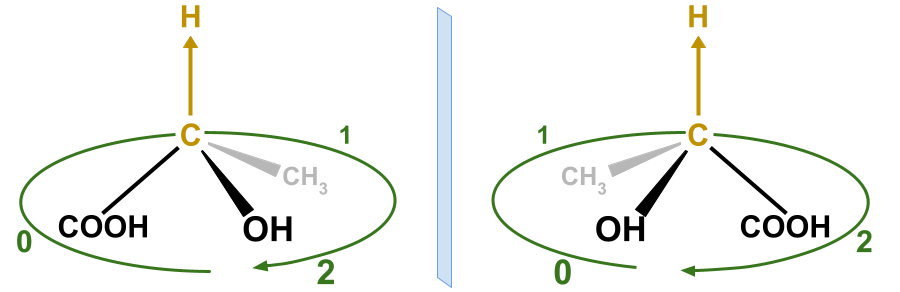
Graph Neural Networks (GNNs) play a fundamental role in many deep learning problems, in particular in cheminformatics. However, typical GNNs cannot capture the concept of chirality, which means they do not distinguish between the 3D graph of a chemical compound and its mirror image (enantiomer). The ability to distinguish between enantiomers is important especially in drug discovery because enantiomers can have very distinct biochemical properties. In this paper, we propose a theoretically justified message-passing scheme, which makes GNNs sensitive to the order of node neighbors. We apply that general concept in the context of molecular chirality to construct Chiral Edge Neural Network (ChiENN) layer which can be appended to any GNN model to enable chirality-awareness. Our experiments show that adding ChiENN layers to a GNN outperforms current state-of-the-art methods in chiral-sensitive molecular property prediction tasks.
PDF Abstract