DAGGER: A sequential algorithm for FDR control on DAGs
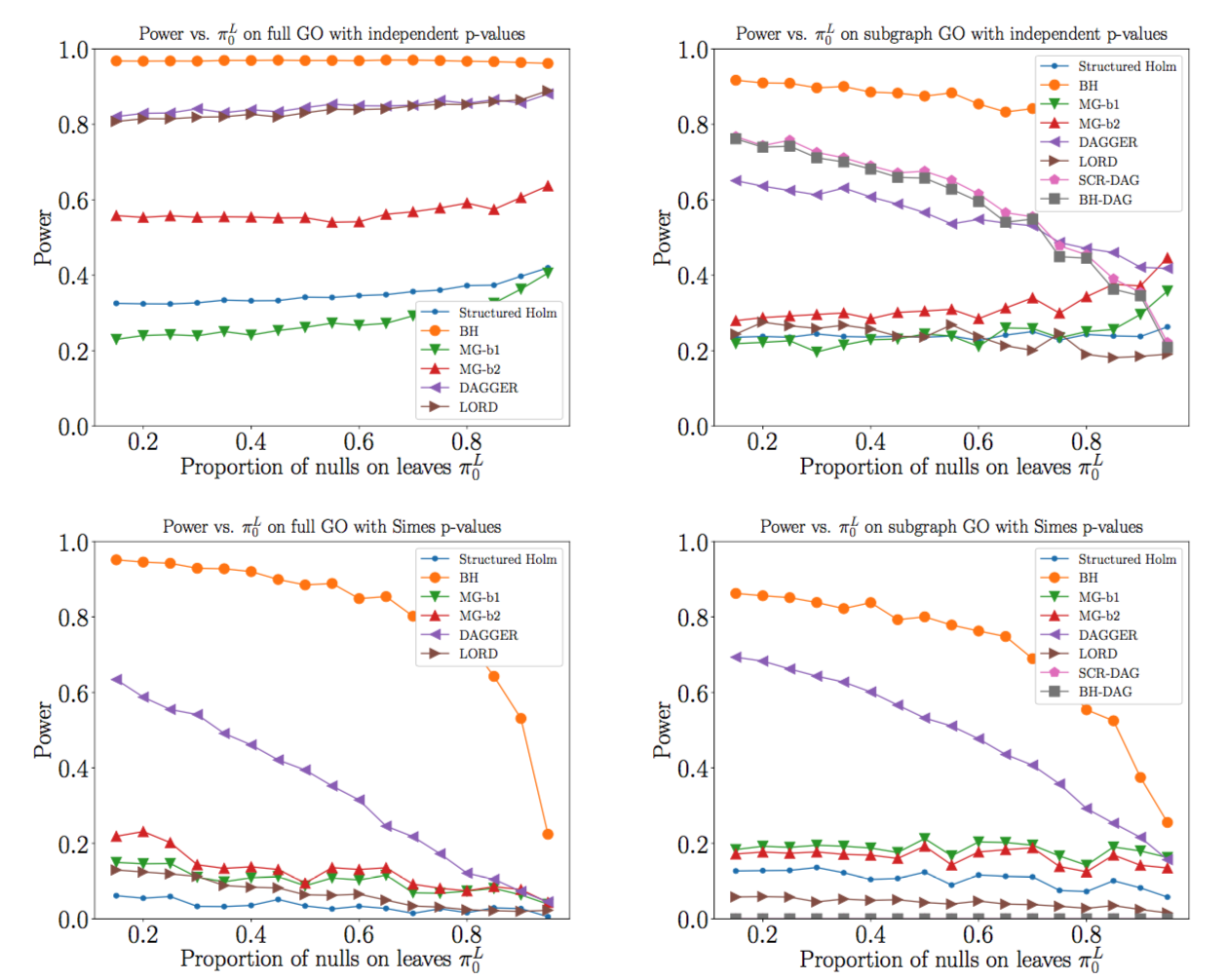
We propose a linear-time, single-pass, top-down algorithm for multiple testing on directed acyclic graphs (DAGs), where nodes represent hypotheses and edges specify a partial ordering in which hypotheses must be tested. The procedure is guaranteed to reject a sub-DAG with bounded false discovery rate (FDR) while satisfying the logical constraint that a rejected node's parents must also be rejected. It is designed for sequential testing settings, when the DAG structure is known a priori, but the $p$-values are obtained selectively (such as in a sequence of experiments), but the algorithm is also applicable in non-sequential settings when all $p$-values can be calculated in advance (such as variable/model selection). Our DAGGER algorithm, shorthand for Greedily Evolving Rejections on DAGs, provably controls the false discovery rate under independence, positive dependence or arbitrary dependence of the $p$-values. The DAGGER procedure specializes to known algorithms in the special cases of trees and line graphs, and simplifies to the classical Benjamini-Hochberg procedure when the DAG has no edges. We explore the empirical performance of DAGGER using simulations, as well as a real dataset corresponding to a gene ontology, showing favorable performance in terms of time and power.
PDF Abstract