DynGFN: Towards Bayesian Inference of Gene Regulatory Networks with GFlowNets
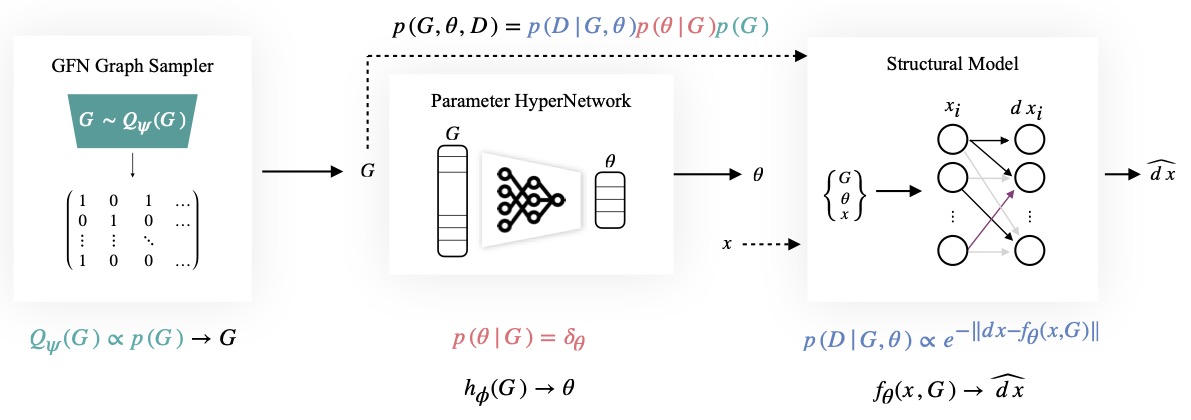
One of the grand challenges of cell biology is inferring the gene regulatory network (GRN) which describes interactions between genes and their products that control gene expression and cellular function. We can treat this as a causal discovery problem but with two non-standard challenges: (1) regulatory networks are inherently cyclic so we should not model a GRN as a directed acyclic graph (DAG), and (2) observations have significant measurement noise, so for typical sample sizes there will always be a large equivalence class of graphs that are likely given the data, and we want methods that capture this uncertainty. Existing methods either focus on challenge (1), identifying cyclic structure from dynamics, or on challenge (2) learning complex Bayesian posteriors over DAGs, but not both. In this paper we leverage the fact that it is possible to estimate the "velocity" of gene expression with RNA velocity techniques to develop an approach that addresses both challenges. Because we have access to velocity information, we can treat the Bayesian structure learning problem as a problem of sparse identification of a dynamical system, capturing cyclic feedback loops through time. Since our objective is to model uncertainty over discrete structures, we leverage Generative Flow Networks (GFlowNets) to estimate the posterior distribution over the combinatorial space of possible sparse dependencies. Our results indicate that our method learns posteriors that better encapsulate the distributions of cyclic structures compared to counterpart state-of-the-art Bayesian structure learning approaches.
PDF Abstract NeurIPS 2023 PDF NeurIPS 2023 Abstract