Explainable Automatic Hypothesis Generation via High-order Graph Walks
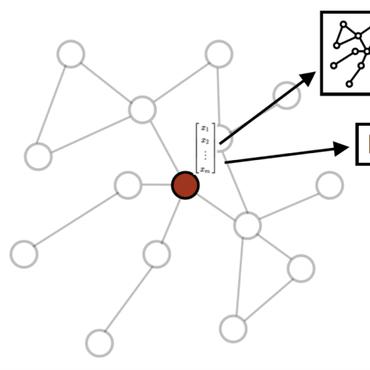
In this paper, we study the automatic hypothesis generation (HG) problem, focusing on explainability. Given pairs of biomedical terms, we focus on link prediction to explain how the prediction was made. This more transparent process encourages trust in the biomedical community for automatic hypothesis generation systems. We use a reinforcement learning strategy to formulate the HG problem as a guided node-pair embedding-based link prediction problem via a directed graph walk. Given nodes in a node-pair, the model starts a graph walk, simultaneously aggregating information from the visited nodes and their neighbors for an improved node-pair representation. Then at the end of the walk, it infers the probability of a link from the gathered information. This guided walk framework allows for explainability via the walk trajectory information. By evaluating our model on predicting the links between millions of biomedical terms in both transductive and inductive settings, we verified the effectiveness of our proposed model on obtaining higher prediction accuracy than baselines and understanding the reason for a link prediction.
PDF Abstract