GIT-Mol: A Multi-modal Large Language Model for Molecular Science with Graph, Image, and Text
Large language models have made significant strides in natural language processing, enabling innovative applications in molecular science by processing textual representations of molecules. However, most existing language models cannot capture the rich information with complex molecular structures or images. In this paper, we introduce GIT-Mol, a multi-modal large language model that integrates the Graph, Image, and Text information. To facilitate the integration of multi-modal molecular data, we propose GIT-Former, a novel architecture that is capable of aligning all modalities into a unified latent space. We achieve a 5%-10% accuracy increase in properties prediction and a 20.2% boost in molecule generation validity compared to the baselines. With the any-to-language molecular translation strategy, our model has the potential to perform more downstream tasks, such as compound name recognition and chemical reaction prediction.
PDF Abstract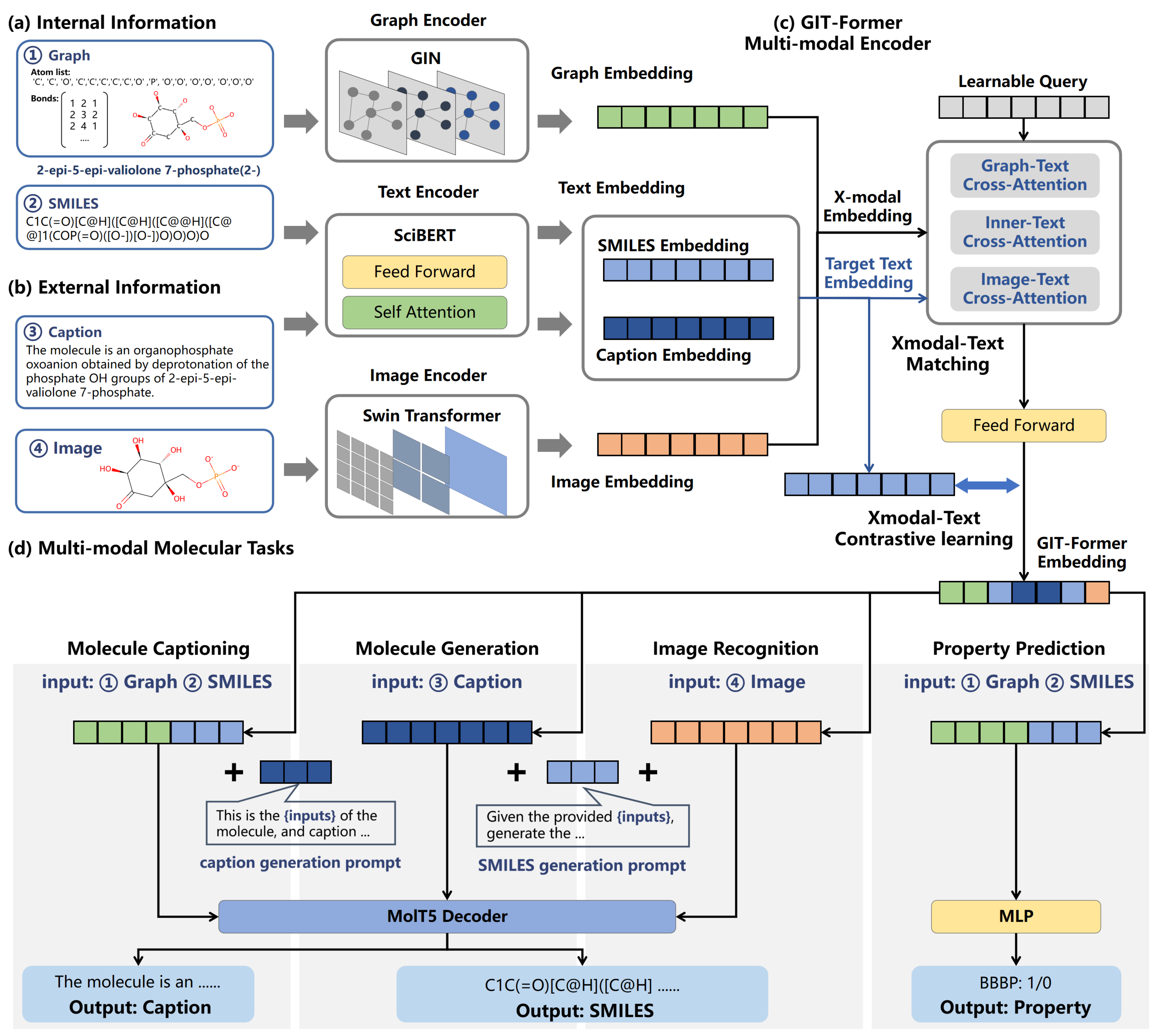













 MoleculeNet
MoleculeNet
 Tox21
Tox21
 ChEBI-20
ChEBI-20
 SIDER
SIDER
