Global Contrast Masked Autoencoders Are Powerful Pathological Representation Learners
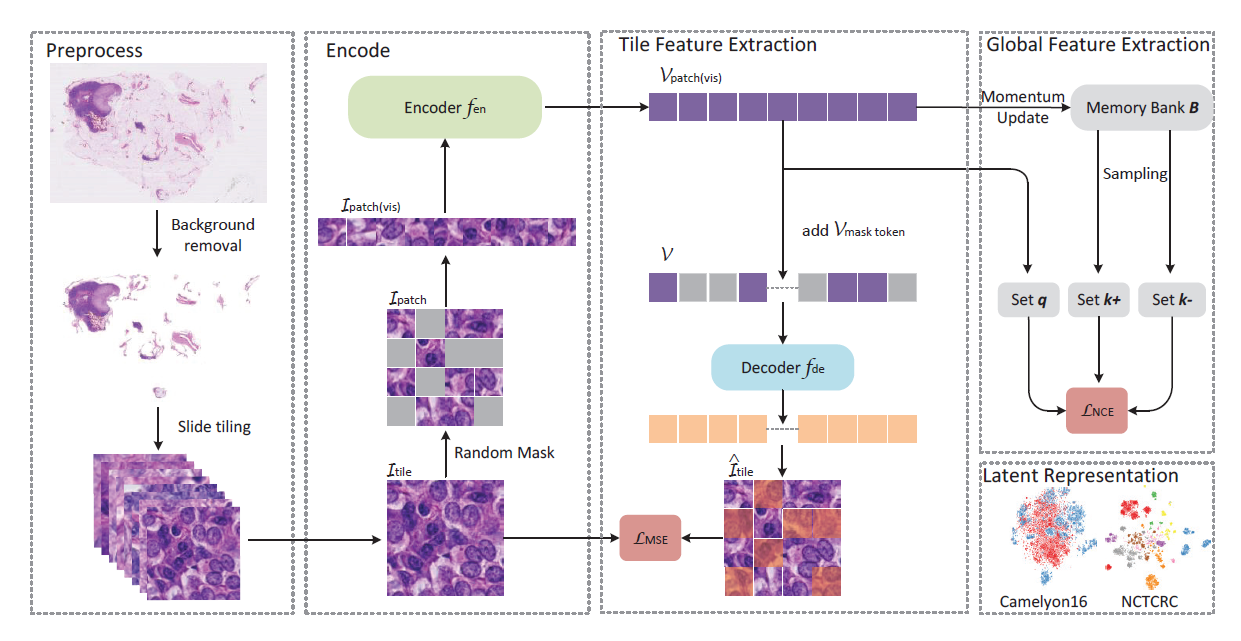
Based on digital pathology slice scanning technology, artificial intelligence algorithms represented by deep learning have achieved remarkable results in the field of computational pathology. Compared to other medical images, pathology images are more difficult to annotate, and thus, there is an extreme lack of available datasets for conducting supervised learning to train robust deep learning models. In this paper, we propose a self-supervised learning (SSL) model, the global contrast-masked autoencoder (GCMAE), which can train the encoder to have the ability to represent local-global features of pathological images, also significantly improve the performance of transfer learning across data sets. In this study, the ability of the GCMAE to learn migratable representations was demonstrated through extensive experiments using a total of three different disease-specific hematoxylin and eosin (HE)-stained pathology datasets: Camelyon16, NCTCRC and BreakHis. In addition, this study designed an effective automated pathology diagnosis process based on the GCMAE for clinical applications. The source code of this paper is publicly available at https://github.com/StarUniversus/gcmae.
PDF Abstract