Improving Biomedical Pretrained Language Models with Knowledge
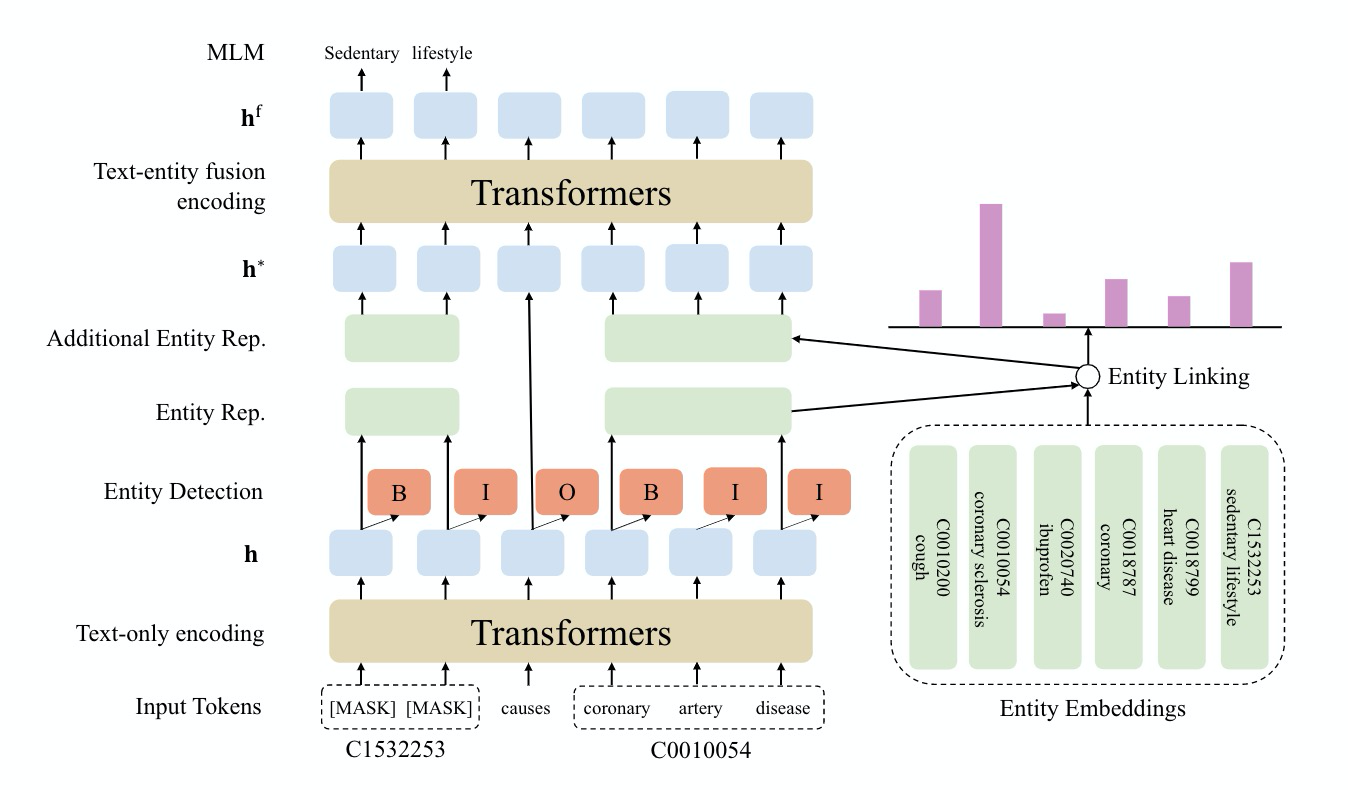
Pretrained language models have shown success in many natural language processing tasks. Many works explore incorporating knowledge into language models. In the biomedical domain, experts have taken decades of effort on building large-scale knowledge bases. For example, the Unified Medical Language System (UMLS) contains millions of entities with their synonyms and defines hundreds of relations among entities. Leveraging this knowledge can benefit a variety of downstream tasks such as named entity recognition and relation extraction. To this end, we propose KeBioLM, a biomedical pretrained language model that explicitly leverages knowledge from the UMLS knowledge bases. Specifically, we extract entities from PubMed abstracts and link them to UMLS. We then train a knowledge-aware language model that firstly applies a text-only encoding layer to learn entity representation and applies a text-entity fusion encoding to aggregate entity representation. Besides, we add two training objectives as entity detection and entity linking. Experiments on the named entity recognition and relation extraction from the BLURB benchmark demonstrate the effectiveness of our approach. Further analysis on a collected probing dataset shows that our model has better ability to model medical knowledge.
PDF Abstract NAACL (BioNLP) 2021 PDF NAACL (BioNLP) 2021 AbstractCode
| Task | Dataset | Model | Metric Name | Metric Value | Global Rank | Benchmark |
|---|---|---|---|---|---|---|
| Named Entity Recognition (NER) | BC2GM | KeBioLM | F1 | 85.1 | # 9 | |
| Named Entity Recognition (NER) | BC5CDR-chemical | KeBioLM | F1 | 93.3 | # 11 | |
| Named Entity Recognition (NER) | BC5CDR-disease | KeBioLM | F1 | 86.1 | # 6 | |
| Relation Extraction | ChemProt | KeBioLM | F1 | 77.5 | # 5 | |
| Relation Extraction | DDI | KeBioLM | F1 | 81.9 | # 2 | |
| Relation Extraction | GAD | KeBioLM | F1 | 84.3 | # 2 | |
| Named Entity Recognition (NER) | JNLPBA | KeBioLM | F1 | 82.0 | # 1 | |
| Named Entity Recognition (NER) | NCBI-disease | KeBioLM | F1 | 89.1 | # 9 |













 BC5CDR
BC5CDR
 DDI
DDI
