PathoDuet: Foundation Models for Pathological Slide Analysis of H&E and IHC Stains
Large amounts of digitized histopathological data display a promising future for developing pathological foundation models via self-supervised learning methods. Foundation models pretrained with these methods serve as a good basis for downstream tasks. However, the gap between natural and histopathological images hinders the direct application of existing methods. In this work, we present PathoDuet, a series of pretrained models on histopathological images, and a new self-supervised learning framework in histopathology. The framework is featured by a newly-introduced pretext token and later task raisers to explicitly utilize certain relations between images, like multiple magnifications and multiple stains. Based on this, two pretext tasks, cross-scale positioning and cross-stain transferring, are designed to pretrain the model on Hematoxylin and Eosin (H\&E) images and transfer the model to immunohistochemistry (IHC) images, respectively. To validate the efficacy of our models, we evaluate the performance over a wide variety of downstream tasks, including patch-level colorectal cancer subtyping and whole slide image (WSI)-level classification in H\&E field, together with expression level prediction of IHC marker and tumor identification in IHC field. The experimental results show the superiority of our models over most tasks and the efficacy of proposed pretext tasks. The codes and models are available at https://github.com/openmedlab/PathoDuet.
PDF Abstract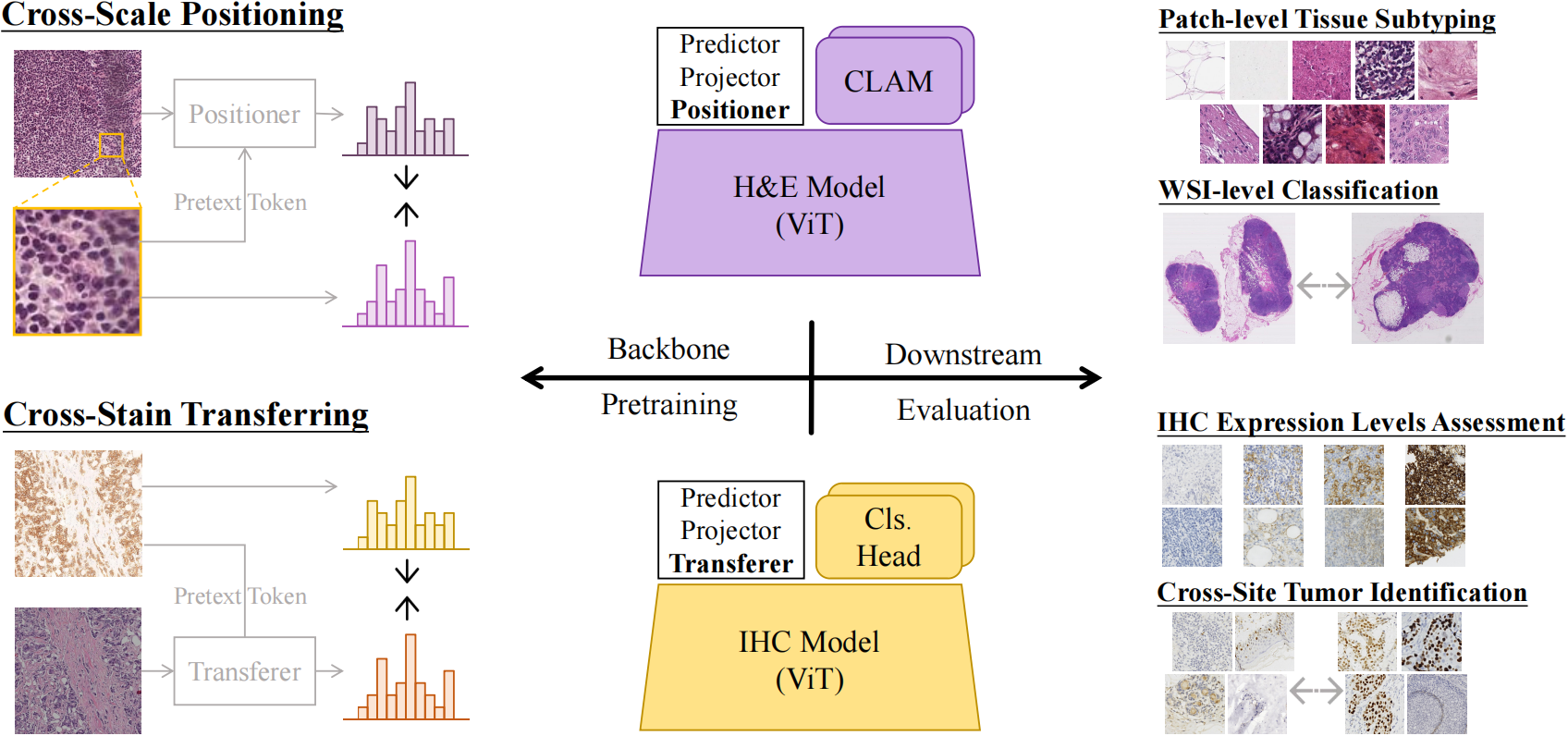


 ImageNet
ImageNet
 BCI
BCI