RandStainNA: Learning Stain-Agnostic Features from Histology Slides by Bridging Stain Augmentation and Normalization
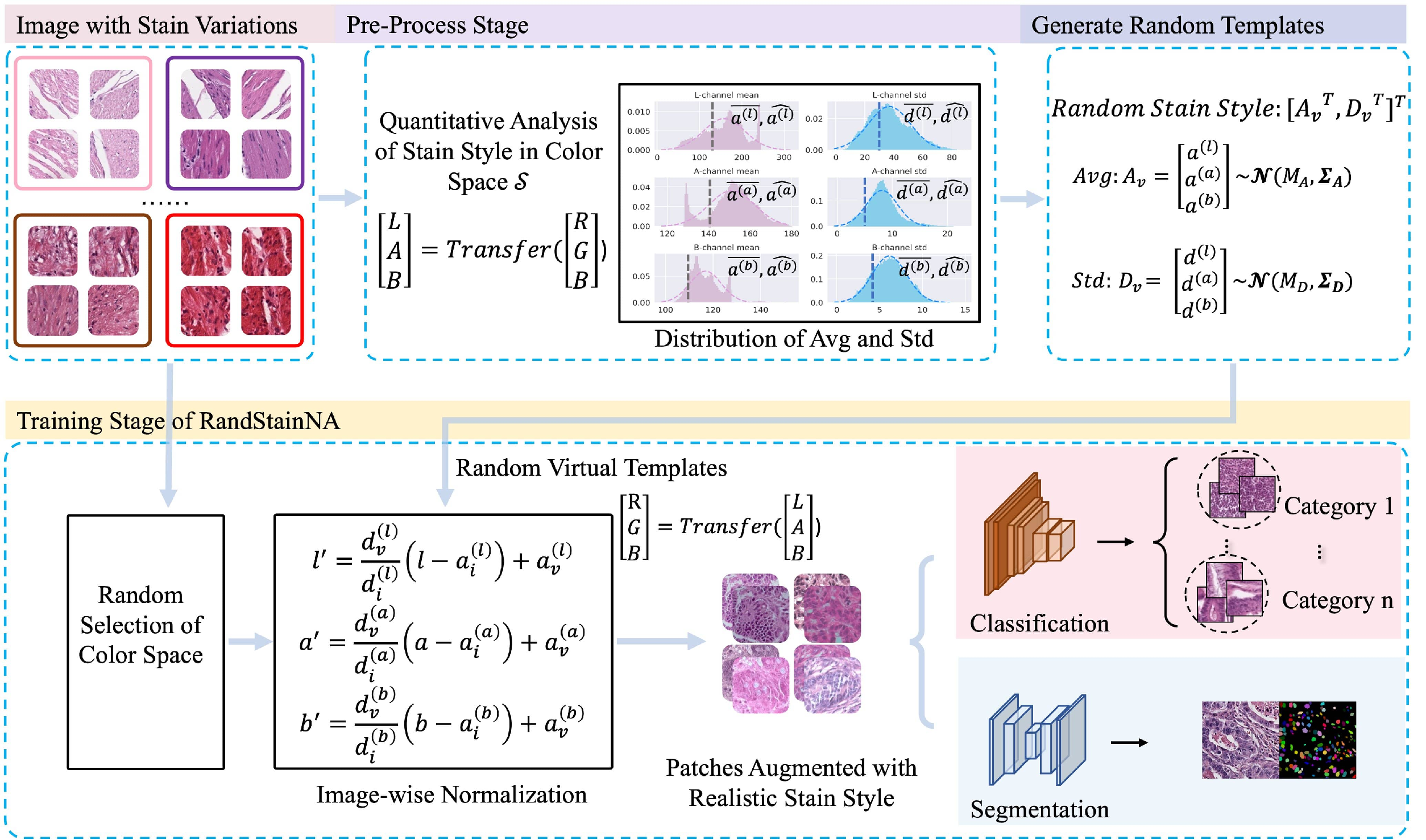
Stain variations often decrease the generalization ability of deep learning based approaches in digital histopathology analysis. Two separate proposals, namely stain normalization (SN) and stain augmentation (SA), have been spotlighted to reduce the generalization error, where the former alleviates the stain shift across different medical centers using template image and the latter enriches the accessible stain styles by the simulation of more stain variations. However, their applications are bounded by the selection of template images and the construction of unrealistic styles. To address the problems, we unify SN and SA with a novel RandStainNA scheme, which constrains variable stain styles in a practicable range to train a stain agnostic deep learning model. The RandStainNA is applicable to stain normalization in a collection of color spaces i.e. HED, HSV, LAB. Additionally, we propose a random color space selection scheme to gain extra performance improvement. We evaluate our method by two diagnostic tasks i.e. tissue subtype classification and nuclei segmentation, with various network backbones. The performance superiority over both SA and SN yields that the proposed RandStainNA can consistently improve the generalization ability, that our models can cope with more incoming clinical datasets with unpredicted stain styles. The codes is available at https://github.com/yiqings/RandStainNA.
PDF Abstract