RetinaRegNet: A Versatile Approach for Retinal Image Registration
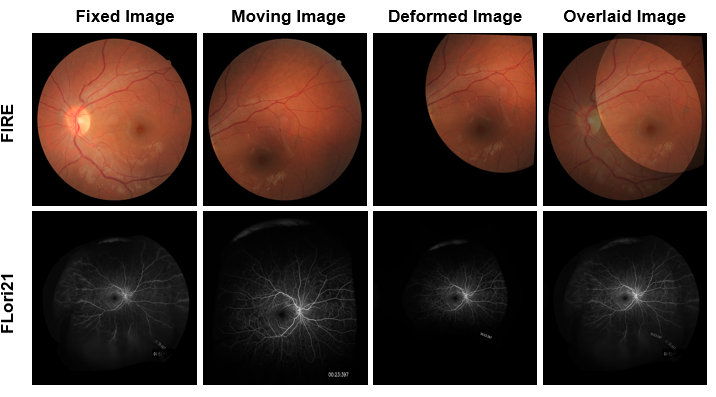
We introduce the RetinaRegNet model, which can achieve state-of-the-art performance across various retinal image registration tasks. RetinaRegNet does not require training on any retinal images. It begins by establishing point correspondences between two retinal images using image features derived from diffusion models. This process involves the selection of feature points from the moving image using the SIFT algorithm alongside random point sampling. For each selected feature point, a 2D correlation map is computed by assessing the similarity between the feature vector at that point and the feature vectors of all pixels in the fixed image. The pixel with the highest similarity score in the correlation map corresponds to the feature point in the moving image. To remove outliers in the estimated point correspondences, we first applied an inverse consistency constraint, followed by a transformation-based outlier detector. This method proved to outperform the widely used random sample consensus (RANSAC) outlier detector by a significant margin. To handle large deformations, we utilized a two-stage image registration framework. A homography transformation was used in the first stage and a more accurate third-order polynomial transformation was used in the second stage. The model's effectiveness was demonstrated across three retinal image datasets: color fundus images, fluorescein angiography images, and laser speckle flowgraphy images. RetinaRegNet outperformed current state-of-the-art methods in all three datasets. It was especially effective for registering image pairs with large displacement and scaling deformations. This innovation holds promise for various applications in retinal image analysis. Our code is publicly available at https://github.com/mirthAI/RetinaRegNet.
PDF Abstract