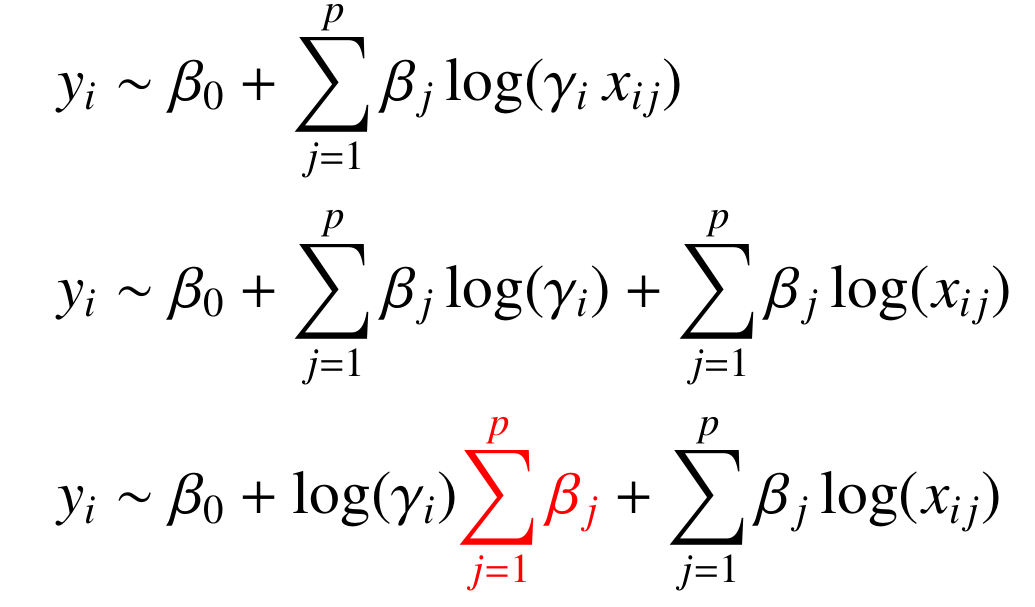Scale-invariant biomarker discovery in urine and plasma metabolite fingerprints
Motivation: Metabolomics data is typically scaled to a common reference like a constant volume of body fluid, a constant creatinine level, or a constant area under the spectrum. Such normalization of the data, however, may affect the selection of biomarkers and the biological interpretation of results in unforeseen ways. Results: First, we study how the outcome of hypothesis tests for differential metabolite concentration is affected by the choice of scale. Furthermore, we observe this interdependence also for different classification approaches. Second, to overcome this problem and establish a scale-invariant biomarker discovery algorithm, we extend linear zero-sum regression to the logistic regression framework and show in two applications to ${}^1$H NMR-based metabolomics data how this approach overcomes the scaling problem. Availability: Logistic zero-sum regression is available as an R package as well as a high-performance computing implementation that can be downloaded at https://github.com/rehbergT/zeroSum
PDF Abstract