Mapping the landscape of histomorphological cancer phenotypes using self-supervised learning on unlabeled, unannotated pathology slides
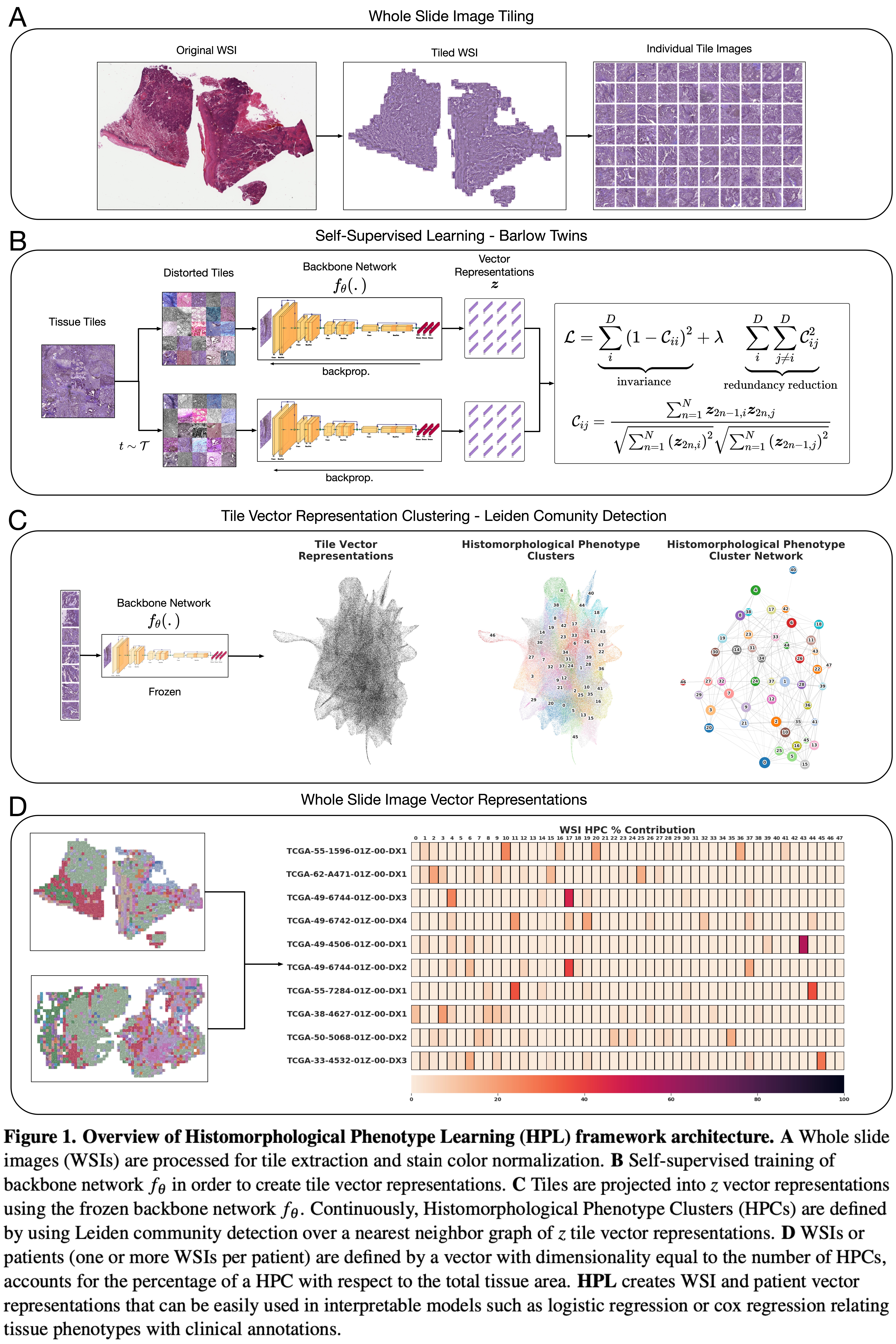
Definitive cancer diagnosis and management depend upon the extraction of information from microscopy images by pathologists. These images contain complex information requiring time-consuming expert human interpretation that is prone to human bias. Supervised deep learning approaches have proven powerful for classification tasks, but they are inherently limited by the cost and quality of annotations used for training these models. To address this limitation of supervised methods, we developed Histomorphological Phenotype Learning (HPL), a fully blue{self-}supervised methodology that requires no expert labels or annotations and operates via the automatic discovery of discriminatory image features in small image tiles. Tiles are grouped into morphologically similar clusters which constitute a library of histomorphological phenotypes, revealing trajectories from benign to malignant tissue via inflammatory and reactive phenotypes. These clusters have distinct features which can be identified using orthogonal methods, linking histologic, molecular and clinical phenotypes. Applied to lung cancer tissues, we show that they align closely with patient survival, with histopathologically recognised tumor types and growth patterns, and with transcriptomic measures of immunophenotype. We then demonstrate that these properties are maintained in a multi-cancer study. These results show the clusters represent recurrent host responses and modes of tumor growth emerging under natural selection. Code, pre-trained models, learned embeddings, and documentation are available to the community at https://github.com/AdalbertoCq/Histomorphological-Phenotype-Learning
PDF Abstract