Trans-Encoder: Unsupervised sentence-pair modelling through self- and mutual-distillations
In NLP, a large volume of tasks involve pairwise comparison between two sequences (e.g. sentence similarity and paraphrase identification). Predominantly, two formulations are used for sentence-pair tasks: bi-encoders and cross-encoders. Bi-encoders produce fixed-dimensional sentence representations and are computationally efficient, however, they usually underperform cross-encoders. Cross-encoders can leverage their attention heads to exploit inter-sentence interactions for better performance but they require task fine-tuning and are computationally more expensive. In this paper, we present a completely unsupervised sentence representation model termed as Trans-Encoder that combines the two learning paradigms into an iterative joint framework to simultaneously learn enhanced bi- and cross-encoders. Specifically, on top of a pre-trained Language Model (PLM), we start with converting it to an unsupervised bi-encoder, and then alternate between the bi- and cross-encoder task formulations. In each alternation, one task formulation will produce pseudo-labels which are used as learning signals for the other task formulation. We then propose an extension to conduct such self-distillation approach on multiple PLMs in parallel and use the average of their pseudo-labels for mutual-distillation. Trans-Encoder creates, to the best of our knowledge, the first completely unsupervised cross-encoder and also a state-of-the-art unsupervised bi-encoder for sentence similarity. Both the bi-encoder and cross-encoder formulations of Trans-Encoder outperform recently proposed state-of-the-art unsupervised sentence encoders such as Mirror-BERT and SimCSE by up to 5% on the sentence similarity benchmarks.
PDF Abstract ICLR 2022 PDF ICLR 2022 Abstract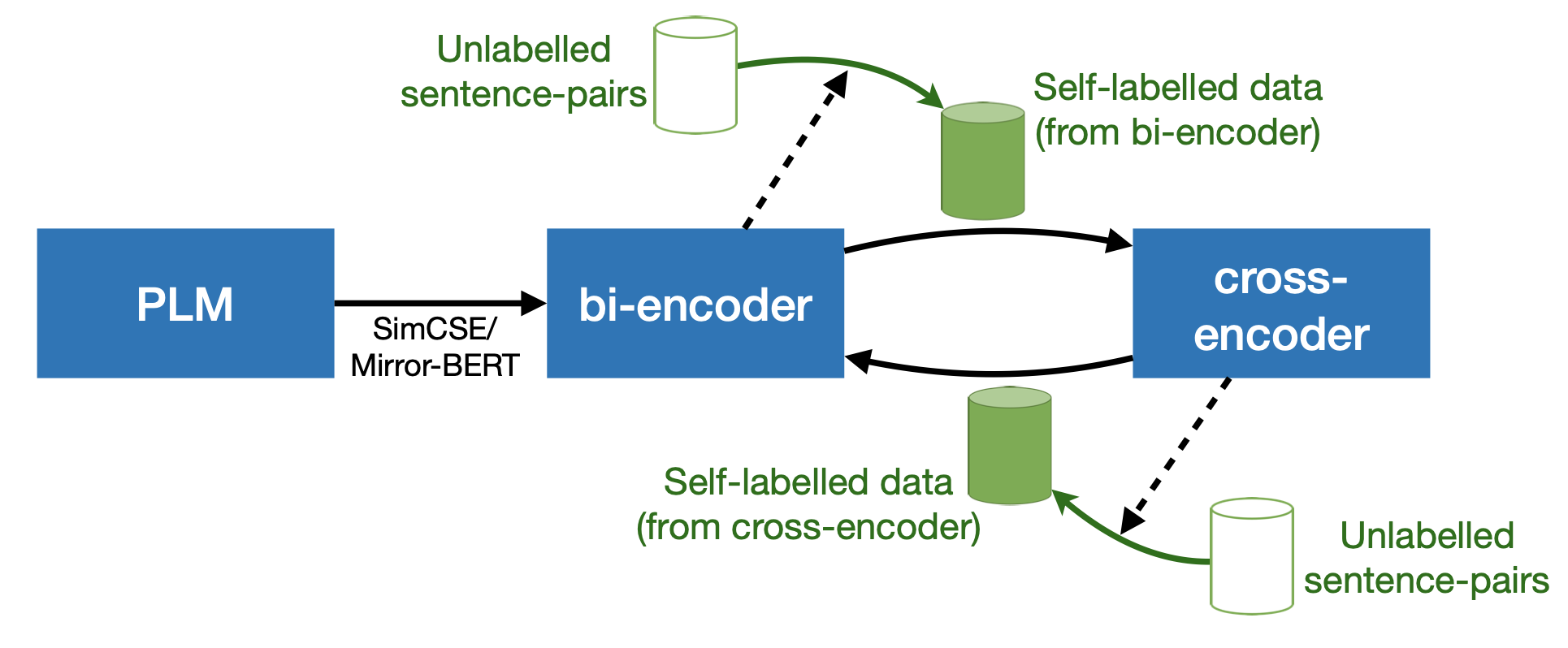













 GLUE
GLUE
 SNLI
SNLI
 QNLI
QNLI
 SICK
SICK
 Quora Question Pairs
Quora Question Pairs
